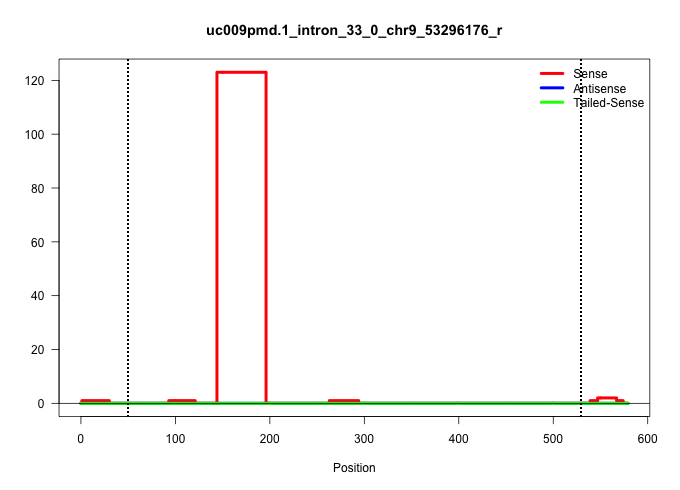
| Gene: Atm | ID: uc009pmd.1_intron_33_0_chr9_53296176_r | SPECIES: mm9 |
 |
|
(1) PIWI.mut |
(8) TESTES |
| TCGTTGGCACACTTATTCCCCTTGTGGATTATCAGGAAGTTCAAGAACAGGTAATTTCCCAGTGCATCTACAACAGAGTATTTGAGACGAAAGATACTCTTAGGAACTGAATGCTTTAAGTTTACCTTAATGAAAATGCTTTTTGATAAATGGTGGAGAAGACATTTTTAGCAAATTAATCCTTGAAATCAATTACTACCCCTTGGTGGTTTTGATCTAATAAATCCAATAGTTTCATCTGTTACACTGAACTGTAGTCAGAAGTAGAGCTAACAGTTCCCATCGTGATGGTCCGTGAGATCTCCTATAAATAACATTCCAGTATGAATTCCTTATGCCTCCTTGTTCAGCACTGTAAGTATTTTAGAAGTGTTTGAAGCCATAACCACTAATAGCTGGGTAATACATATGCCGTTAGAACATGTCTTCTGGACTAGGGTAGCAGAGGTTTGTTTTTATAATGTCATAATGGGATTTGGCTCTTTAAATATAATTAAGACTTCTCTGTCTTCGTTTGATAATTGTCTAGGTATTGGACCTGTTGAAGTACTTAGTGATAGATAACAAAGACAATAAAAA |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................................................................GATAAATGGTGGAGAAGACATTTTTAGCAAATTAATCCTTGAAATCAATTAC............................................................................................................................................................................................................................................................................................................................................................................................... | 52 | 1 | 123.00 | 123.00 | 123.00 | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................TACTTAGTGATAGATAACAAAGACAAT..... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - |
| .CGTTGGCACACTTATTCCCCTTGTGGATT..................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - |
| .......................................................................................................................................................................................................................................................................GTAGAGCTAACAGTTCCCATCGTGATGGTCC............................................................................................................................................................................................................................................................................................. | 31 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - |
| .............................................................................................ATACTCTTAGGAACTGAATGCTTTAAGT.......................................................................................................................................................................................................................................................................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - |
| ...........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................TGTTGAAGTACTTAGTGATAGATAACAA............ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - |
| TCGTTGGCACACTTATTCCCCTTGTGGATTATCAGGAAGTTCAAGAACAGGTAATTTCCCAGTGCATCTACAACAGAGTATTTGAGACGAAAGATACTCTTAGGAACTGAATGCTTTAAGTTTACCTTAATGAAAATGCTTTTTGATAAATGGTGGAGAAGACATTTTTAGCAAATTAATCCTTGAAATCAATTACTACCCCTTGGTGGTTTTGATCTAATAAATCCAATAGTTTCATCTGTTACACTGAACTGTAGTCAGAAGTAGAGCTAACAGTTCCCATCGTGATGGTCCGTGAGATCTCCTATAAATAACATTCCAGTATGAATTCCTTATGCCTCCTTGTTCAGCACTGTAAGTATTTTAGAAGTGTTTGAAGCCATAACCACTAATAGCTGGGTAATACATATGCCGTTAGAACATGTCTTCTGGACTAGGGTAGCAGAGGTTTGTTTTTATAATGTCATAATGGGATTTGGCTCTTTAAATATAATTAAGACTTCTCTGTCTTCGTTTGATAATTGTCTAGGTATTGGACCTGTTGAAGTACTTAGTGATAGATAACAAAGACAATAAAAA |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................................................................................................TGGTTTTGATCTAATAAAg................................................................................................................................................................................................................................................................................................................................................................... | 19 | g | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 |
| ....................................................................................................................................................................................................................................................................................................................................................................................AAGCCATAACCACTtgga............................................................................................................................................................................................. | 18 | tgga | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - |
| ..........................................................................................................................................................................................................TGGTTTTGATCTAgtcg........................................................................................................................................................................................................................................................................................................................................................................ | 17 | gtcg | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - |