
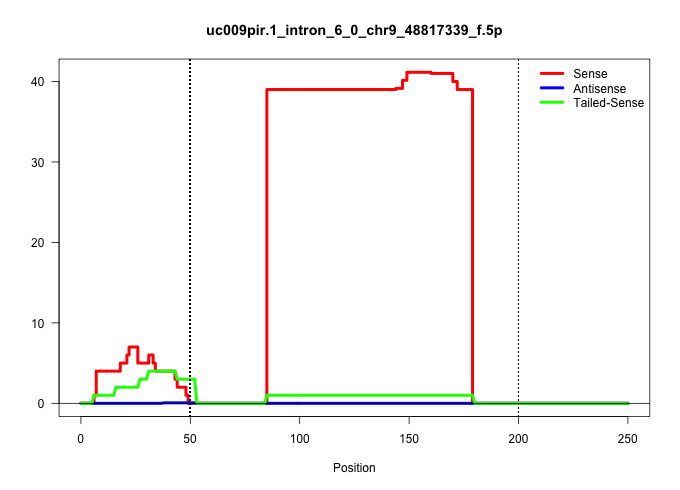
| Gene: Usp28 | ID: uc009pir.1_intron_6_0_chr9_48817339_f.5p | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(1) PIWI.ip |
(1) PIWI.mut |
(12) TESTES |
| CAGCCCTGGATCTACTGAAGGGAGCCTTCCGATCATCCGAGGAGCAGCAGGTACTTTGTTCCCTTGTCATATTTCAAAGGCCAGTGTCTGAGTCACCCTTTGCTAGTGTGGTACTGCTATTTGAATTGTTACTGTATGACAAGCTCTGAACAAAGACAAGATGACTGCCTTCATCTGCCCTTTATTTCGCTCTCAGAAGGTGTAGTTAGAGCAGGTGTGTTCACATGGGTGAGAGGTGCACGATGCGCTG .....................................................................................................(((..(((.((((.((..........))....)))).))))))...(((.((((...((((((......))))))...))))..))).............................................................. ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | mjTestesWT3() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................GTCTGAGTCACCCTTTGCTAGTGTGGTACTGCTATTTGAATTGTTACTGTAT................................................................................................................. | 52 | 1 | 43.00 | 43.00 | 43.00 | - | - | - | - | - | - | - | - | - | - | - |
| .......GGATCTACTGAAGGGAGCC................................................................................................................................................................................................................................ | 19 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - |
| ........................................................................................................AGTGTGGTACTGCTATT................................................................................................................................. | 17 | 2 | 1.50 | 1.50 | - | - | - | 1.50 | - | - | - | - | - | - | - | - |
| ......................AGCCTTCCGATCATCCGAGGAGCAGC.......................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ....................GGAGCCTTCCGATCATCCGAGGAGCAGCAG........................................................................................................................................................................................................ | 30 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................ACAAAGACAAGATGACTGCCT................................................................................ | 21 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................GAACAAAGACAAGATGACTGCCTTC.............................................................................. | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...........................TCCGATCATCCGAGGAGCAGCAGcaa..................................................................................................................................................................................................... | 26 | caa | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...............................ATCATCCGAGGAGCAGCAGcag..................................................................................................................................................................................................... | 22 | cag | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..................................ATCCGAGGAGCAGCAGcat..................................................................................................................................................................................................... | 19 | cat | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...............................ATCATCCGAGGAGCAGCA......................................................................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ......TGGATCTACTGAAGGGAGCCTTCCGctc........................................................................................................................................................................................................................ | 28 | ctc | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .......GGATCTACTGAAGGGAGCCTTCCGATC........................................................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..................AGGGAGCCTTCCGATCATCCGAGGAG.............................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ......TGGATCTACTGAAGGGAGCCTTCCGAT......................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ................GAAGGGAGCCTTCCGATCATCCGAGGAa.............................................................................................................................................................................................................. | 28 | a | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .....................GAGCCTTCCGATCATCCGAGGA............................................................................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ................................................................................................................................................TCTGAACAAAGACAAG.......................................................................................... | 16 | 7 | 0.14 | 0.14 | - | - | - | - | - | - | - | - | - | - | 0.14 | - |
| CAGCCCTGGATCTACTGAAGGGAGCCTTCCGATCATCCGAGGAGCAGCAGGTACTTTGTTCCCTTGTCATATTTCAAAGGCCAGTGTCTGAGTCACCCTTTGCTAGTGTGGTACTGCTATTTGAATTGTTACTGTATGACAAGCTCTGAACAAAGACAAGATGACTGCCTTCATCTGCCCTTTATTTCGCTCTCAGAAGGTGTAGTTAGAGCAGGTGTGTTCACATGGGTGAGAGGTGCACGATGCGCTG .....................................................................................................(((..(((.((((.((..........))....)))).))))))...(((.((((...((((((......))))))...))))..))).............................................................. ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | mjTestesWT3() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................GAGGAGCAGCAGGTA..................................................................................................................................................................................................... | 15 | 16 | 0.06 | 0.06 | - | - | - | - | - | - | - | - | - | - | - | 0.06 |