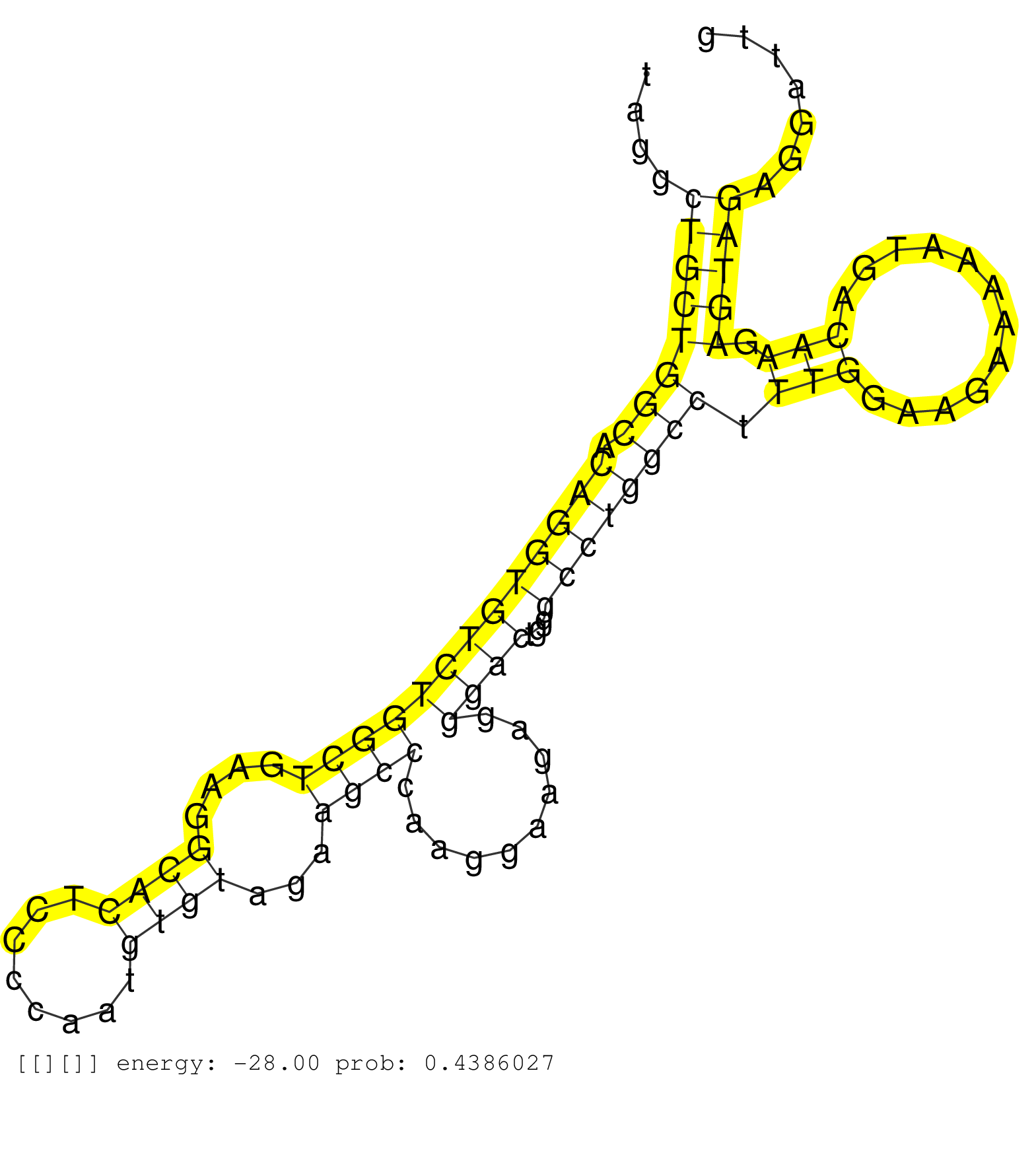

| Gene: Dscaml1 | ID: uc009pfz.1_intron_7_0_chr9_45482807_f.5p | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(1) OVARY |
(4) PIWI.ip |
(1) PIWI.mut |
(15) TESTES |
| CATCCAGCCACAGCTTTCCATCAGCCAGAGTGTCCACGTGGCCGTCAAAGGTAGGTCCCTCCCCGCCTGCAGCCTGCCTGCCTCCTAGGCTGCTGGCACAGGTGTCTGGCTGAAGGCACTCCCCAATGTGTAGAAGCCCAAGGAAGAGGGACTGGGCCTGGCCTTTGGAAGAAAAATGACAAGAGTAGAGGATTGGGTTGGACACAAGGCTAAAAGCTCCAGGGAAAAAGACAACCAGCACTGACCTTGG .........................................................................................((((((((.(((((((((((((....((((........))))...))))..........))))...)))))))).(((............))).))))).............................................................. .....................................................................................86...........................................................................................................195..................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | mjTestesWT3() Testes Data. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................................TTGGAAGAAAAATGACAAGAGTAGAGG........................................................... | 27 | 1 | 5.00 | 5.00 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................TGGAAGAAAAATGACAAGAGTAGAGGA.......................................................... | 27 | 1 | 3.00 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................TGGAAGAAAAATGACAAGAGTAGAGG........................................................... | 26 | 1 | 3.00 | 3.00 | - | 1.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................GAGGATTGGGTTGGACACAAGGCTA...................................... | 25 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................TGACAAGAGTAGAGGATTGGGTTGG................................................. | 25 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................TTGGAAGAAAAATGACAAGAGTAGAGGA.......................................................... | 28 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................TGACAAGAGTAGAGGATTGGGTTGGA................................................ | 26 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ....................................................................................................................................................................TTGGAAGAAAAATGACAAGAGTAGAG............................................................ | 26 | 1 | 2.00 | 2.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................AAATGACAAGAGTAGAGGATTGGGTT................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................TGGAAGAAAAATGACAAGAGTAGAGGt.......................................................... | 27 | t | 1.00 | 3.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................TGGAAGAAAAATGACAAGAGTAGAGGAT......................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................TAAAAGCTCCAGGGAAAAAGACAAC............... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..........................................................................................TGCTGGCACAGGTGTCTGGCTGAAGGCACTCC................................................................................................................................ | 32 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................TGTGTAGAAGCCCAAGGAAGAGGGA................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................TAAAAGCTCCAGGGAAAAAGACAACC.............. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..............................................................................................................................................................................AATGACAAGAGTAGAGGATTGGGTTG.................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................TGTGTAGAAGCCCAAGGAAGAGGGACTG................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................TGTGTAGAAGCCCAAGGAAGAGGGACT................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .................................................................................................................................................................................GACAAGAGTAGAGGATTGGGTTGGACACA............................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................TTTGGAAGAAAAATGACAAGAGTAGA............................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................AGAGGATTGGGTTGGA................................................ | 16 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | 0.17 | - | - | - | - | - | - | - |
| CATCCAGCCACAGCTTTCCATCAGCCAGAGTGTCCACGTGGCCGTCAAAGGTAGGTCCCTCCCCGCCTGCAGCCTGCCTGCCTCCTAGGCTGCTGGCACAGGTGTCTGGCTGAAGGCACTCCCCAATGTGTAGAAGCCCAAGGAAGAGGGACTGGGCCTGGCCTTTGGAAGAAAAATGACAAGAGTAGAGGATTGGGTTGGACACAAGGCTAAAAGCTCCAGGGAAAAAGACAACCAGCACTGACCTTGG .........................................................................................((((((((.(((((((((((((....((((........))))...))))..........))))...)))))))).(((............))).))))).............................................................. .....................................................................................86...........................................................................................................195..................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | mjTestesWT3() Testes Data. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................CAGAGTGTCCACGTGGCCGTCAAAGGTA..................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .TCCAGCCACAGCTTTCCAa...................................................................................................................................................................................................................................... | 19 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...................................................................................................................GCACTCCCCAATGTGTAGAAGCCCA.............................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............CATCAGCCAGAGTaaga........................................................................................................................................................................................................................... | 17 | aaga | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...CCAGCCACAGCTTTCCATCAGCCAGA............................................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .ATCCAGCCACAGCTTTCCATCAGCCA............................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................TCCCCGCCTGCAGCC................................................................................................................................................................................ | 15 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 |