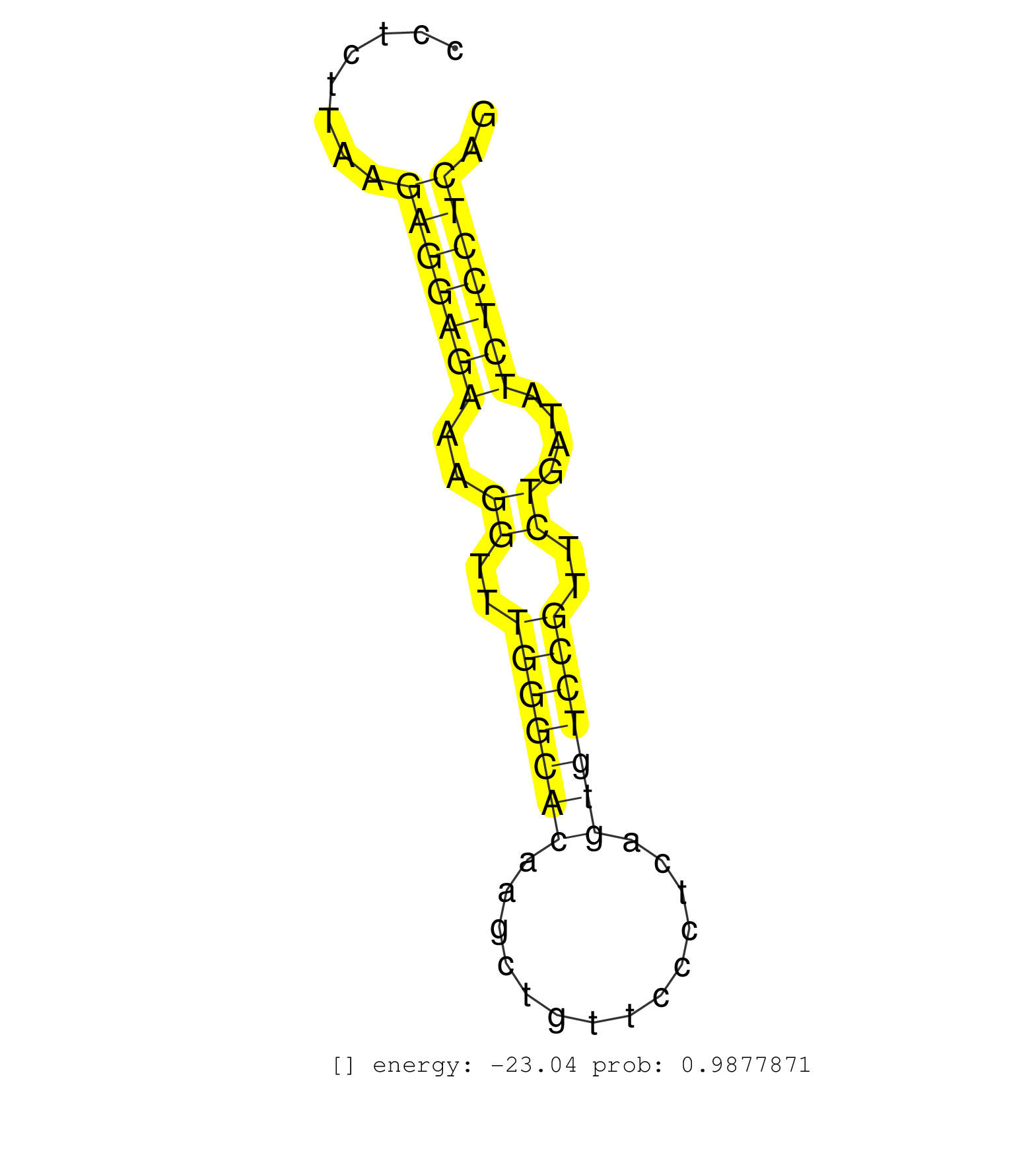
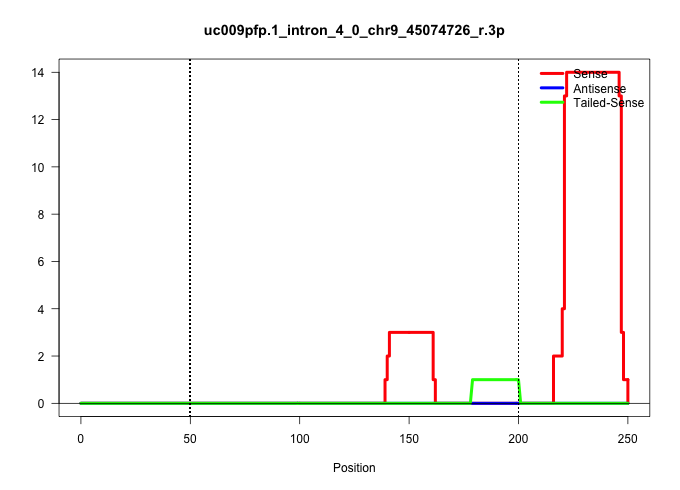
| Gene: Il10ra | ID: uc009pfp.1_intron_4_0_chr9_45074726_r.3p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(4) PIWI.ip |
(2) PIWI.mut |
(14) TESTES |
| TTAGGGGGGAATGAGCTACTTAAATTCCCACCCAGGGGCTTTCTAGAACATGGATGTCAACAGAGACCTTGGCCAAGGGCCTGAGGACTGTTCCCTTTGTCATTAAGGAAACGTTCTGCCTGGCCTTTCCCTCCCCTCTTAAGAGGAGAAAGGTTTGGGCACAAGCTGTTCCCTCAGTGTCCGTTCTGATATCTCCTCAGGTACGGAAACTCAACCTGGAATGACATCCATATCTGTAGAAAGGCTCAGG ..............................................................................................................................................(((((((..((..(((((((..............)))))))..))....))))))).................................................... ......................................................................................................................................135..............................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................................................................................................................TGACATCCATATCTGTAGAAAGGCTC... | 26 | 1 | 7.00 | 7.00 | 3.00 | 2.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - |
| ...........................................................................................................................................TAAGAGGAGAAAGGTTTGGGCA......................................................................................... | 22 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................ATGACATCCATATCTGTAGAAAGGCTC... | 27 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .............................................................................................................................................AGAGGAGAAAGGTTTGGGCAa........................................................................................ | 21 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...................................................................................................................................................................................TCCGTTCTGATATCTCCTCAGt................................................. | 22 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .............................................................................................................................................................................................................................TGACATCCATATCTGTAGAAAGGCTCAGG | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..............................................................................................................................................................................................................................GACATCCATATCTGTAGAAAGGCTCA.. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................AAGAGGAGAAAGGTTTGGGCA......................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................TGACATCCATATCTGTAGAAAGGCTCA.. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .............................................................................................................................................AGAGGAGAAAGGTTTGGGCAC........................................................................................ | 21 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................TGGAATGACATCCATATCTGTAGAAAGGCTC... | 31 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................TGGAATGACATCCATATCTGTAGAAAGGCT.... | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TTAGGGGGGAATGAGCTACTTAAATTCCCACCCAGGGGCTTTCTAGAACATGGATGTCAACAGAGACCTTGGCCAAGGGCCTGAGGACTGTTCCCTTTGTCATTAAGGAAACGTTCTGCCTGGCCTTTCCCTCCCCTCTTAAGAGGAGAAAGGTTTGGGCACAAGCTGTTCCCTCAGTGTCCGTTCTGATATCTCCTCAGGTACGGAAACTCAACCTGGAATGACATCCATATCTGTAGAAAGGCTCAGG ..............................................................................................................................................(((((((..((..(((((((..............)))))))..))....))))))).................................................... ......................................................................................................................................135..............................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................................................................................................AAACTCAACCTGGAAtgtt............................. | 19 | tgtt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |