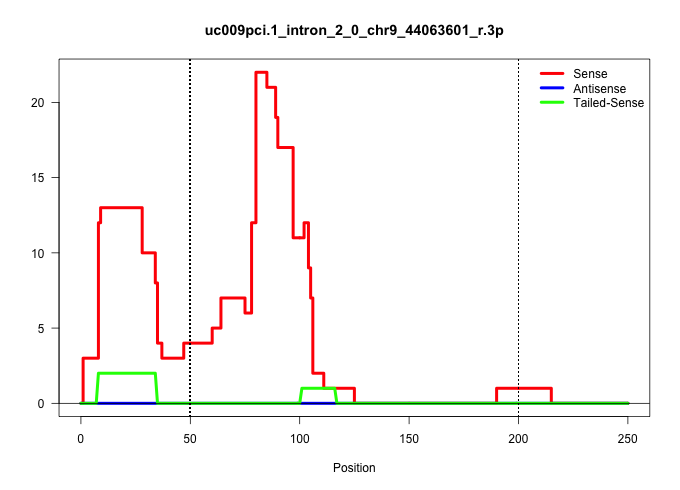
| Gene: Nlrx1 | ID: uc009pci.1_intron_2_0_chr9_44063601_r.3p | SPECIES: mm9 |
 |
|
(1) OTHER.mut |
(3) PIWI.ip |
(2) PIWI.mut |
(13) TESTES |
| ATGTCCCCTGACATAGCAGAGCACAGCAGGGCAGGATGGAGAAGTTCTGTGGGCAGAGGCCACTTGAGCAACAGGGGGCAAGGAATGTGTGGATGGCAGGCAGGAGGCTGGTAAGGAGGGGGCACACTCCATGGGAAGACCCCATGCTGACCTGCTTGCTTGGTCTCTTGCCTCTCCTCTCCGGCCACCTTCTGCTCCAGGTTGCAACTCAACAATCTGGGCCCCGAGGCCTGCAGAGACCTCCGAGACC |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT4() Testes Data. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................CAAGGAATGTGTGGATGGC......................................................................................................................................................... | 19 | 1 | 6.00 | 6.00 | - | 6.00 | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................AGGAATGTGTGGATGGCAGGCAGGAG................................................................................................................................................ | 26 | 1 | 5.00 | 5.00 | 1.00 | - | - | 4.00 | - | - | - | - | - | - | - | - | - |
| ........TGACATAGCAGAGCACAGCAGGGCAGG....................................................................................................................................................................................................................... | 27 | 1 | 4.00 | 4.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - |
| .TGTCCCCTGACATAGCAGAGCACAGCA.............................................................................................................................................................................................................................. | 27 | 1 | 3.00 | 3.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ........TGACATAGCAGAGCACAGCAGGGCAG........................................................................................................................................................................................................................ | 26 | 1 | 3.00 | 3.00 | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | 1.00 |
| ................................................................................AGGAATGTGTGGATGGCAGGCAGG.................................................................................................................................................. | 24 | 1 | 3.00 | 3.00 | 1.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - |
| ................................................................TGAGCAACAGGGGGCAAGGAATGTGT................................................................................................................................................................ | 26 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - |
| ........TGACATAGCAGAGCACAGCAGGGCAGGA...................................................................................................................................................................................................................... | 28 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - |
| ........TGACATAGCAGAGCACAGCAGGGCAGt....................................................................................................................................................................................................................... | 27 | t | 2.00 | 3.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................TGGAGAAGTTCTGTGGGCAGAGGCCACT.......................................................................................................................................................................................... | 28 | 1 | 2.00 | 2.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ................................................................................AGGAATGTGTGGATGGCAGGCAGGA................................................................................................................................................. | 25 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ................................................................TGAGCAACAGGGGGCAAGGAATGTG................................................................................................................................................................. | 25 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ............................................................CACTTGAGCAACAGGGGGCAAGGAA..................................................................................................................................................................... | 25 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - |
| ..............................................................................................................................................................................................TCTGCTCCAGGTTGCAACTCAACAA................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..................................GATGGAGAAGTTCTGTGGGCAGAGGC.............................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .........GACATAGCAGAGCACAGCAGGGCAGGAT..................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...............................................TGTGGGCAGAGGCCACTTGAGCAACAGG............................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ......................................................................................................GGAGGCTGGTAAGGAGGGGGCAC............................................................................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .....................................................................................TGTGTGGATGGCAGGCAGGAGGCTGG........................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .....................................................................................................AGGAGGCTGGTAAaat..................................................................................................................................... | 16 | aat | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ATGTCCCCTGACATAGCAGAGCACAGCAGGGCAGGATGGAGAAGTTCTGTGGGCAGAGGCCACTTGAGCAACAGGGGGCAAGGAATGTGTGGATGGCAGGCAGGAGGCTGGTAAGGAGGGGGCACACTCCATGGGAAGACCCCATGCTGACCTGCTTGCTTGGTCTCTTGCCTCTCCTCTCCGGCCACCTTCTGCTCCAGGTTGCAACTCAACAATCTGGGCCCCGAGGCCTGCAGAGACCTCCGAGACC |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT4() Testes Data. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|