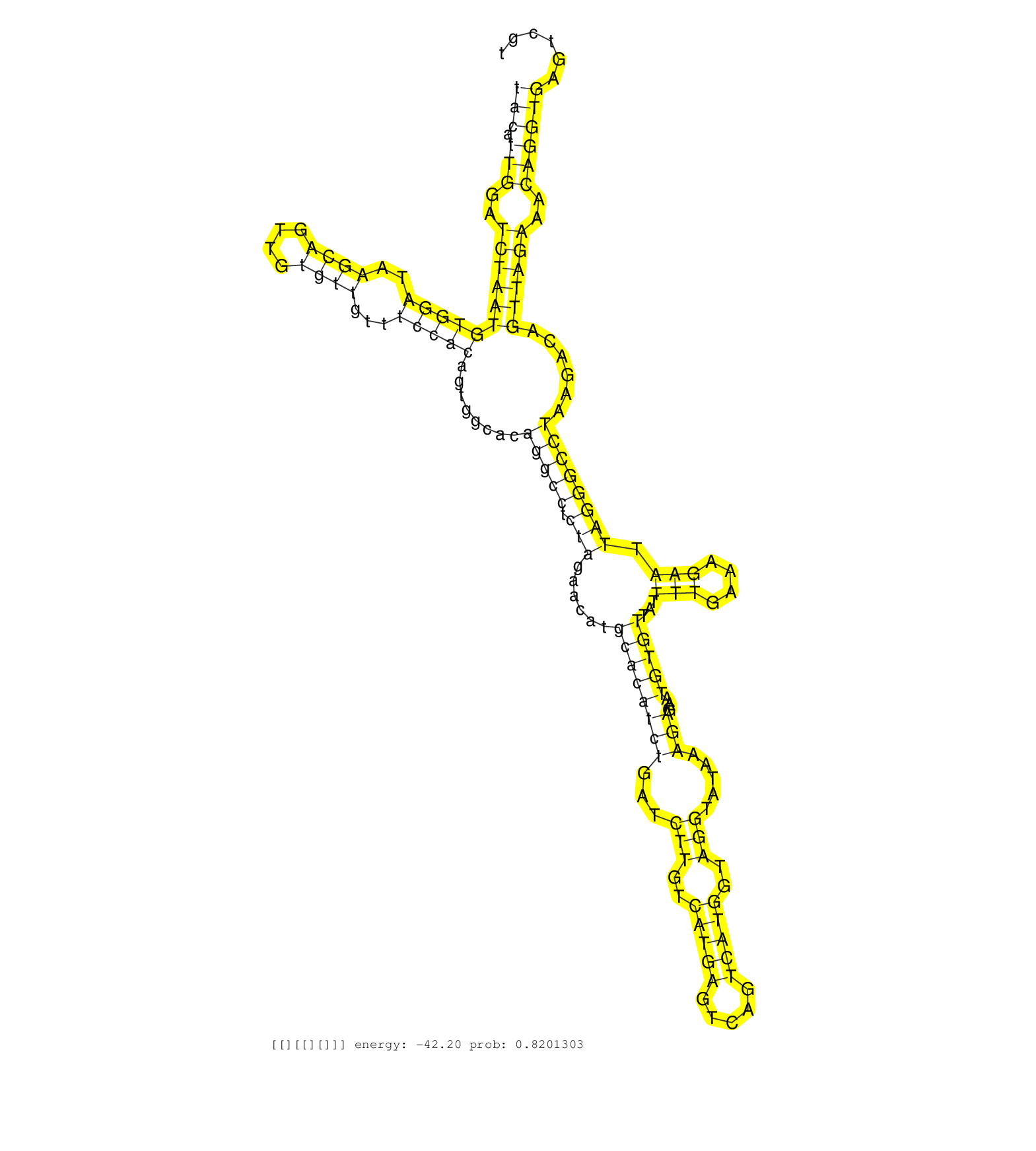

| Gene: Cbl | ID: uc009pce.1_intron_2_0_chr9_43961942_r | SPECIES: mm9 |
 |
 |
|
(7) OTHER.mut |
(6) PIWI.ip |
(29) TESTES |
| GTTCAGAAGCCAGAGCCCAAACGGCCGTTAGAGGCAACCCAGAGTTCACGGTATGCTCACAGCAATCCTATTTGGAGCCAAGTGTCTTTGCTGTCTTTTACCTGCTTGTTTGCTAATTGCCAGAAGTAAAGCAGAAAGAGAAATCTAGACCCCAGTACATTGGATCTAATGTGGATAAGCAGTTGTGTTGTTTCCACAGTGGCACAGGCCTCTAGAACATGCACATCTGATCTTGTCATGAGTCAGTCATGGTAGGTATAAAGAGAATGTGTTATTTTGAAAGAATTAGGGCCTAAGACAGTTAGAAACAGGTGAGTCGTCTTAAATTTGCAAATAACAGAATTTTGTTTGTGAAGAGCATTGTACCTCTTTGTGATTCAGAGCATGTGACTGTGACCAGCAGATCGACAGCTGTACCTATGAAGCGATGT ...........................................................................................................................................................(((.(((..(((((((((((..((((....))))...)))))........(((((.(((......((((((((...(((..(((((.....)))))..))).....)))...)))))...(((....))).))))))))......))))))..))))))..................................................................................................................... ...........................................................................................................................................................156.................................................................................................................................................................320............................................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT4() Testes Data. (testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR363958(GSM822760) Adult-WTSmall RNA Miwi IP. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM475281(GSM475281) total RNA. (testes) | mjTestesWT2() Testes Data. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................................................................................................GATCTTGTCATGAGTCAGTCATGGTAGGTATAAAGAGAATGTGTTATTTTGA....................................................................................................................................................... | 52 | 1 | 119.00 | 119.00 | 119.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................................................................................................................................................................................................TGTGACTGTGACCAGCAGATCGACAGCTG................. | 29 | 1 | 6.00 | 6.00 | - | 2.00 | - | 3.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................................................................................................................................................................................................TGACCAGCAGATCGACAGCTGTACCT............ | 26 | 1 | 6.00 | 6.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .................................................................................................................................................................................................................................................................................................................................................................................................TGTGACTGTGACCAGCAGATCGACAGC................... | 27 | 1 | 5.00 | 5.00 | - | - | - | - | 1.00 | 1.00 | - | - | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................................................................................................................................................................................GCATGTGACTGTGACCAGCAGAT.......................... | 23 | 1 | 4.00 | 4.00 | - | - | 1.00 | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................................................................................................................................................................................................TGTGACTGTGACCAGCAGATCGACAGCT.................. | 28 | 1 | 3.00 | 3.00 | - | - | - | - | 1.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................................................................................................................................................................................................................ACAGCTGTACCTATGAAGCG.... | 20 | 1 | 3.00 | 3.00 | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................................................................................................................................................................................................TGTGACTGTGACCAGCAGATCGACAGCTGT................ | 30 | 1 | 2.00 | 2.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................................................................................................................................................................................................TGACCAGCAGATCGACAGCTGTACCTA........... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................................................................................................................................................................................AGCATGTGACTGTGACCAGCAGA........................... | 23 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................................................................................................................................................................................................TGACCAGCAGATCGACAGCTGTACC............. | 25 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................TGGATCTAATGTGGATAAGCAGTTG...................................................................................................................................................................................................................................................... | 25 | 1 | 2.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................TAGAGGCAACCCAGAGTTCACGagc.......................................................................................................................................................................................................................................................................................................................................................................................... | 25 | agc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................................................................................................................................................................................................GCAGATCGACAGCTGTACCTATGAAGC..... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................................................................................TTTTGAAAGAATTAGGGCCTAAGA..................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................................................................................................................................................................................................ACCAGCAGATCGACAGCTGTACCTATGA........ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................................................................................................................................................................................................TGACCAGCAGATCGACAGCTGTAact............ | 26 | act | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .................................................................................................................................................................................................................................................................................................................................................................................................TGTGACTGTGACCAGCAGATCGACAG.................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................................................................................................................................................................................................TGTGACCAGCAGATCGACAGt................... | 21 | t | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................................................................................................................................................................................................TGACCAGCAGATCGACAGCTGTACCTAT.......... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................................................................................................................................................................................AGCATGTGACTGTGACCAGCAGATCGACAGC................... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................................................................................................................................................................................AGCATGTGACTGTGACCAGCAGATCGAC...................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................................TCTGATCTTGTCATGAGTCAGTCATGGTAt................................................................................................................................................................................ | 30 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................................................................................................................................................................................................TGACTGTGACCAGCAGATCGACAGCTGTAC.............. | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................................................................................................................................................................................................TGTGACCAGCAGATCGACAGCTGTAC.............. | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................................................................................................................................................................................AGCATGTGACTGTGACCAGCAGATCGt....................... | 27 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................................................................................................................................................................................................GCAGATCGACAGCTGTACCTATGAAGttt... | 29 | ttt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................................................................................................................................................................................CATGTGACTGTGACCAGCAGATCGACA..................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................................................................................................................................................................................................TGACCAGCAGATCGACAGCTGTACCTATGt........ | 30 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .............................................................................................................................................................................................................................................................................................................................................................................................AGCATGTGACTGTGACCAGC.............................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................................................................................................................................................................................AGCATGTGACTGTGACCAGCAGATCGACA..................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................................................................................................................................................................................GCATGTGACTGTGACCAGCAGATCGA....................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................................................................................................................................................................................................TGACTGTGACCAGCAGATCGACAGCT.................. | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................................................................................................................................................................................AGCATGTGACTGTGACCAGCAGATCGA....................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................................................................................................................................................................................................TGACCAGCAGATCGACAGCTG................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ............................TAGAGGCAACCCAGAGTTCACGagcc......................................................................................................................................................................................................................................................................................................................................................................................... | 26 | agcc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................TACATTGGATCTAATGTGGATAAGCAGgtt...................................................................................................................................................................................................................................................... | 30 | gtt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .........................................................................................................................................................................................................................................................................................................................................................................................................TGACCAGCAGATCGACAGCTGTACCcatg......... | 29 | catg | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .................................................................................................................................................................................................................................................................................................................................................................................................TGTGACTGTGACCAGCAGA........................... | 19 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 |
| ..............................................................................................................................................................................................................................................................................................................................................................................................GCATGTGACTGTGACC................................. | 16 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - | - | - | - |
| GTTCAGAAGCCAGAGCCCAAACGGCCGTTAGAGGCAACCCAGAGTTCACGGTATGCTCACAGCAATCCTATTTGGAGCCAAGTGTCTTTGCTGTCTTTTACCTGCTTGTTTGCTAATTGCCAGAAGTAAAGCAGAAAGAGAAATCTAGACCCCAGTACATTGGATCTAATGTGGATAAGCAGTTGTGTTGTTTCCACAGTGGCACAGGCCTCTAGAACATGCACATCTGATCTTGTCATGAGTCAGTCATGGTAGGTATAAAGAGAATGTGTTATTTTGAAAGAATTAGGGCCTAAGACAGTTAGAAACAGGTGAGTCGTCTTAAATTTGCAAATAACAGAATTTTGTTTGTGAAGAGCATTGTACCTCTTTGTGATTCAGAGCATGTGACTGTGACCAGCAGATCGACAGCTGTACCTATGAAGCGATGT ...........................................................................................................................................................(((.(((..(((((((((((..((((....))))...)))))........(((((.(((......((((((((...(((..(((((.....)))))..))).....)))...)))))...(((....))).))))))))......))))))..))))))..................................................................................................................... ...........................................................................................................................................................156.................................................................................................................................................................320............................................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT3() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT4() Testes Data. (testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR363958(GSM822760) Adult-WTSmall RNA Miwi IP. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM475281(GSM475281) total RNA. (testes) | mjTestesWT2() Testes Data. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) |
|---|