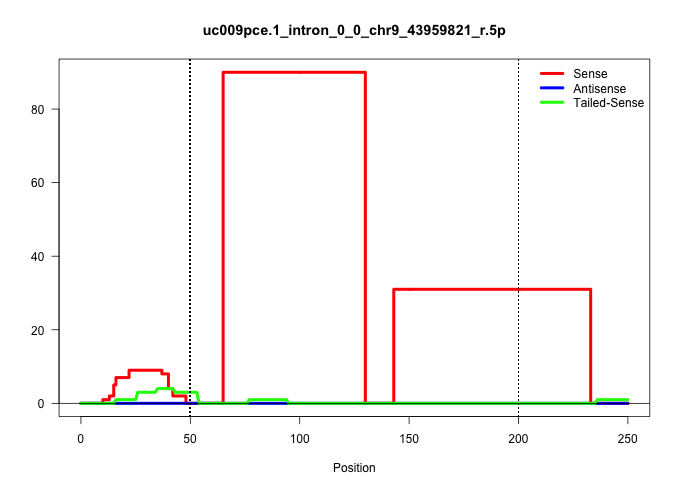
| Gene: Cbl | ID: uc009pce.1_intron_0_0_chr9_43959821_r.5p | SPECIES: mm9 |
 |
 |
|
(4) OTHER.mut |
(2) PIWI.ip |
(14) TESTES |
| CAATGCCAGCTCCTCCTTTGGCTGGTTGTCTTTGGATGGTGACCCTACAAGTGAGTCTCCAGGCTGTCTTAGCTAGTCCTGGATTGTTGTCTGGCCTCTGGTTTATAACTGAAAGTCATGACTTTGTCTCGTGTCAATAAGTCAGTGAGTAGAAGTAGTGGACTGTGTCCTGGATCTATATGTTGGAAAGCTGGTCCTAGGTCCTACATTAGGAAGTGACACTCTGAAGATGGTGCTTAAACCACAGCTC .....................................................................................................(((((.(((((......((((..........)))).....)))))..)))))......((((...)))).(((((.....(((....))).)))))..................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT3() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................GTCTTAGCTAGTCCTGGATTGTTGTCTGGCCTCTGGTTTATAACTGAAAGTC..................................................................................................................................... | 52 | 1 | 90.00 | 90.00 | 90.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................AGTGAGTAGAAGTAGTGGACTGTGTCCTGGATCTATATGTTGGAAAGCTGGT....................................................... | 52 | 1 | 31.00 | 31.00 | 31.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................AAGTGACACTCTGAAGATGG................. | 20 | 1 | 4.00 | 4.00 | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............CTTTGGCTGGTTGTCTTTGGATGGT.................................................................................................................................................................................................................. | 25 | 1 | 3.00 | 3.00 | - | - | 2.00 | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ......................TGGTTGTCTTTGGATGGTGACCCTAC.......................................................................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - |
| ................TTTGGCTGGTTGTCTTTGGATGGTGA................................................................................................................................................................................................................ | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - |
| ..........................TGTCTTTGGATGGTGACCCTACAAactt.................................................................................................................................................................................................... | 28 | actt | 2.00 | 0.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - |
| .............................................................................CCTGGATTGTTGTCcaaa........................................................................................................................................................... | 18 | caaa | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................GAAGTGACACTCTGAAGATGGTGCTTAAACCACA.... | 34 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ............................................................................................................................................................................................................................................TTAAACCACAGCTCcatt | 18 | catt | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ................TTTGGCTGGTTGTCTTTGGATGGcgac............................................................................................................................................................................................................... | 27 | cgac | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...................................ATGGTGACCCTACAAactt.................................................................................................................................................................................................... | 19 | actt | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..........TCCTCCTTTGGCTGGTTGTCTTTGGAT..................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .............TCCTTTGGCTGGTTGTCTTTGGATGGT.................................................................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TATGTTGGAAAGCTGG........................................................ | 16 | 4 | 0.25 | 0.25 | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - |
| CAATGCCAGCTCCTCCTTTGGCTGGTTGTCTTTGGATGGTGACCCTACAAGTGAGTCTCCAGGCTGTCTTAGCTAGTCCTGGATTGTTGTCTGGCCTCTGGTTTATAACTGAAAGTCATGACTTTGTCTCGTGTCAATAAGTCAGTGAGTAGAAGTAGTGGACTGTGTCCTGGATCTATATGTTGGAAAGCTGGTCCTAGGTCCTACATTAGGAAGTGACACTCTGAAGATGGTGCTTAAACCACAGCTC .....................................................................................................(((((.(((((......((((..........)))).....)))))..)))))......((((...)))).(((((.....(((....))).)))))..................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT3() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................TTGTTGTCTGGCCTCaatt........................................................................................................................................................ | 19 | aatt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ................................................................................TTGTTGTCTGGCCTCTatt....................................................................................................................................................... | 19 | att | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |