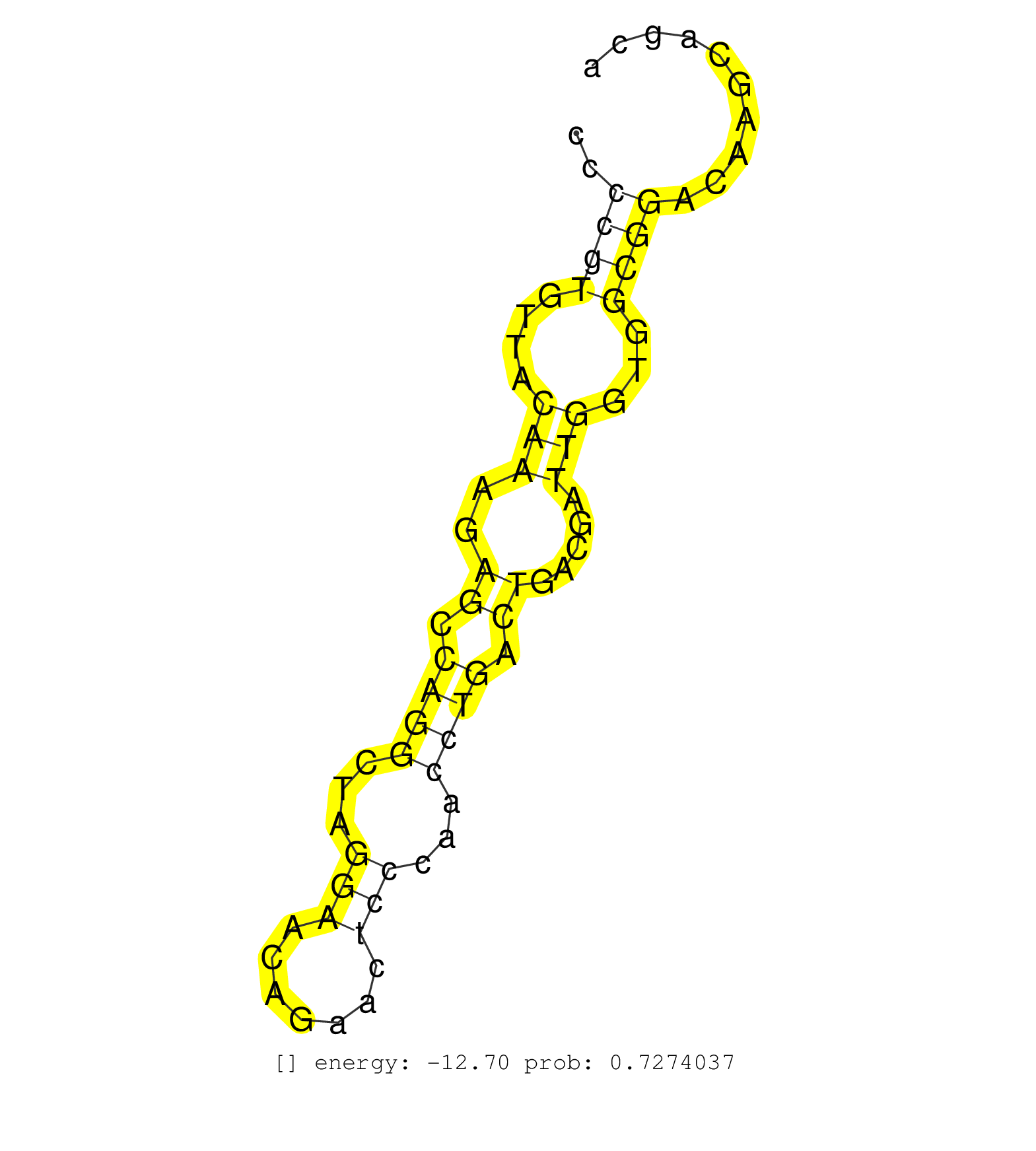
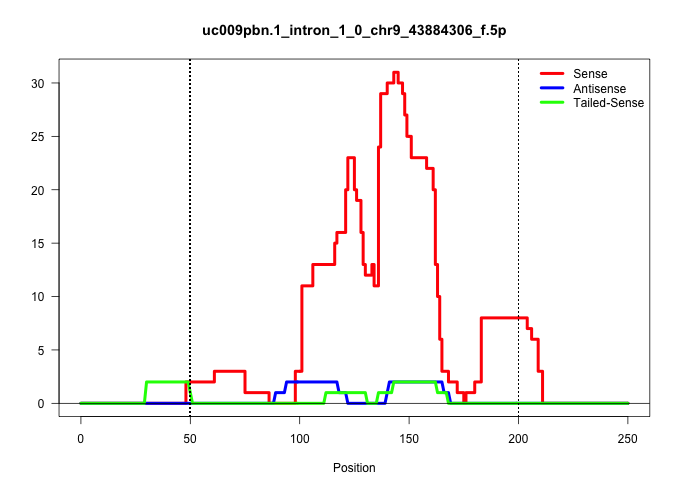
| Gene: Usp2 | ID: uc009pbn.1_intron_1_0_chr9_43884306_f.5p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.ip |
(5) OTHER.mut |
(4) PIWI.ip |
(1) PIWI.mut |
(23) TESTES |
| GCCAGGCCTCTGGGCCCAGCCGCTCCAGTTCTCCAGGGCGAGACACCATGGTGAGTTTACTCTTGGGGATCCAGTGAGGGACTGGGGAGACAGCCCCGTGTTACAAAGAGCCAGGCTAGGAACAGAACTCCCAACCTGACTGACGATTGGTGGCGGACAAGCAGCAGAGATCATCTTCCACGATGATCAAAGCATGACTTGGTGGCTCCATGGCTCAGAGCCATGGGGAAGGGGCTGGCCCGTGCTTCAG ...............................................................................................((((....(((..((.((((...(((.......)))...)))).)).....)))...)))).............................................................................................. .............................................................................................94......................................................................166.................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................................TGACTGACGATTGGTGGCGGACAAGC........................................................................................ | 26 | 1 | 6.00 | 6.00 | - | 1.00 | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .......................................................................................................................................................................................TGATCAAAGCATGACTTGGTGGCTCC......................................... | 26 | 1 | 3.00 | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - |
| ..................................................................................................TGTTACAAAGAGCCAGGCTAGGAACAG............................................................................................................................. | 27 | 1 | 3.00 | 3.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................TACAAAGAGCCAGGCTAGGAACAGAAC.......................................................................................................................... | 27 | 1 | 3.00 | 3.00 | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................TGATCAAAGCATGACTTGGTGGCTCCAT....................................... | 28 | 1 | 3.00 | 3.00 | - | - | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................TACAAAGAGCCAGGCTAGGAACAGAACT......................................................................................................................... | 28 | 1 | 3.00 | 3.00 | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................GACTGACGATTGGTGGCGGACAAGCAG...................................................................................... | 27 | 1 | 3.00 | 3.00 | - | 1.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................TGACTGACGATTGGTGGCGGACAAG......................................................................................... | 25 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................TGGTGAGTTTACTCTTGGGGATCCAGT............................................................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................AGAGCCAGGCTAGGAACAGAACTCCCAA.................................................................................................................... | 28 | 1 | 2.00 | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................ACAGAACTCCCAACCTGACTGACGATTGGT................................................................................................... | 30 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................CAGAACTCCCAACCTGACTGACGATTG..................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................TGACTGACGATTGGTGGCGGACAAGCA....................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................TGACTGACGATTGGTGGCGGACAAGCAGC..................................................................................... | 29 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................TAGGAACAGAACTCCCAACCTGACTGA........................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................CGATGATCAAAGCATGACTTGGTGGC............................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................CGATTGGTGGCGGACAAGCAGCAGAGATC.............................................................................. | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................CGATTGGTGGCGGACAAGCAGCcga.................................................................................. | 25 | cga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................AGGAACAGAACTCCCAACCTGACTGACGA........................................................................................................ | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................ACCTGACTGACGATTGGTGGCGGAC............................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................TAGGAACAGAACTCCCAACCTGACTGACG......................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................TCCACGATGATCAAAGCATGACTTGGTG.............................................. | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................CTCCAGGGCGAGACACCATa........................................................................................................................................................................................................ | 20 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................ACAGAACTCCCAACCTGACTGACGATT...................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................CAGAACTCCCAACCTGACTGACGATT...................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................CTTGGGGATCCAGTGAGGGACTGGG.................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................AGGCTAGGAACAGAAaaac....................................................................................................................... | 19 | aaac | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ........................................................................................................................................TGACTGACGATTGGTGGCGGACAAGCAG...................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................TTGGTGGCGGACAAGCAGCAGAGATCATC........................................................................... | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................GACTGACGATTGGTGGCGGACAAGC........................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................GACTGACGATTGGTGGCGGACAAGCA....................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................TGACGATTGGTGGCGGACAAGCAGC..................................................................................... | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................CGATTGGTGGCGGACAAGCAGCAGA.................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................TACAAAGAGCCAGGCTAGGAACAGAACTC........................................................................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................CTCCAGGGCGAGACACCATGa....................................................................................................................................................................................................... | 21 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................TGATCAAAGCATGACTTGGTGGCTCCATG...................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .........................................................................................................................ACAGAACTCCCAACCTGACTGACGAT....................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................TACAAAGAGCCAGGCTAGGAACAGA............................................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................TGACTGACGATTGGTGGCGGACAAGCt....................................................................................... | 27 | t | 1.00 | 6.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| GCCAGGCCTCTGGGCCCAGCCGCTCCAGTTCTCCAGGGCGAGACACCATGGTGAGTTTACTCTTGGGGATCCAGTGAGGGACTGGGGAGACAGCCCCGTGTTACAAAGAGCCAGGCTAGGAACAGAACTCCCAACCTGACTGACGATTGGTGGCGGACAAGCAGCAGAGATCATCTTCCACGATGATCAAAGCATGACTTGGTGGCTCCATGGCTCAGAGCCATGGGGAAGGGGCTGGCCCGTGCTTCAG ...............................................................................................((((....(((..((.((((...(((.......)))...)))).)).....)))...)))).............................................................................................. .............................................................................................94......................................................................166.................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................ACAGAACTCCCAACCTggc................................................................................................................. | 19 | ggc | 4.00 | 0.00 | - | - | - | - | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................AACTCCCAACCTGACTtggc............................................................................................................. | 20 | tggc | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 |
| .............................................................................................................................................GACGATTGGTGGCGGACAAGCAGCAGAG................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..............................................................................................CCCGTGTTACAAAGAGCCAGGCTAGGAA................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................ACAGCCCCGTGTTACAAAGAGCCAGGCTA.................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................TGACGATTGGTGGCGGACAAGCAGCA.................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |