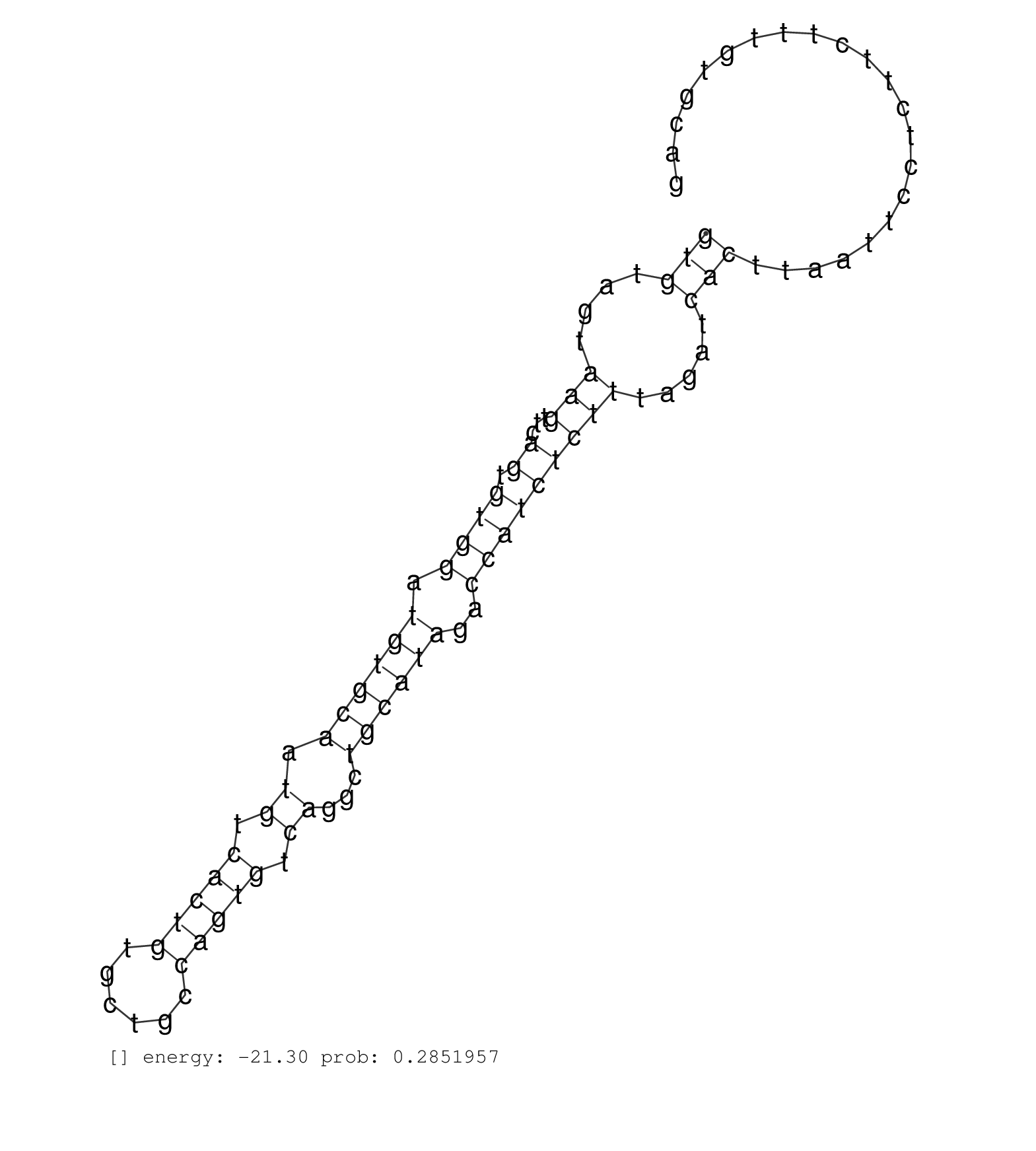

| Gene: Arhgef12 | ID: uc009pba.1_intron_24_0_chr9_42809635_r | SPECIES: mm9 |
 |
 |
|
(6) OTHER.mut |
(2) OVARY |
(2) PIWI.ip |
(1) PIWI.mut |
(18) TESTES |
| GTTTCAGCACCTGAAGGTTCCAGTTCCTGAGGAAATCTCCGTGGATCTAGGTAAGTTTGAAGCACCCGAGTCCCTGAAGATTAGTGTCTCCATCAAACTGTGGCTCGCCTCAGATCCGCTGTTTTATGTTAGTTTAGTCAGGCTACTTTAACCATGTGACTCACAGCTGAAAGAGAGTCTACTACACTGAGAGGATGAAAGGATGAGAGCTGGCTTCTGACAGGAAGTGCAGTAACTGCACTGAGTAAGTTCTCTGGTGTAGTAAGTTCAGTGTGGATGTGCAATGTCACTGTGCTGCCAGTGTCAGGCTGCATAGACCATCTCTTTAGATCACTTAATTCCTCTTCTTTGTGCAGAAAAGAGAAGACCTGAGCTTATTCCTGAGGATCTGCATCGCCTCTACATT ................................................................................................................................................................................................................................................................(((....(((...((.((((.((((((.((.(((((......))))).))...))))))..))))))))).....)))........................................................................ ................................................................................................................................................................................................................................................................257................................................................................................356................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | mjTestesWT1() Testes Data. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR014231(GSM319955) 16.5 dpc total. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................................................................................................TGAAAGGATGAGAGCTGGCTTCTGACA........................................................................................................................................................................................ | 27 | 1 | 28.00 | 28.00 | 6.00 | 4.00 | 4.00 | 4.00 | - | - | 1.00 | 2.00 | 2.00 | 2.00 | - | - | 1.00 | - | - | 1.00 | - | - | - | 1.00 |
| ..................................................................................................................................................................................................................................................................................................................................................................AGAAAAGAGAAGACCTGAGCTTATTCCT........................ | 28 | 1 | 3.00 | 3.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..................................................................................................................................................................................................................................................................................................................................................................AGAAAAGAGAAGACCTGAGCTTATTCCTG....................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................................................GTAAGTTCTCTGGTGTAGTAAGa........................................................................................................................................... | 23 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..................................................GTAAGTTTGAAGCACCCGAGTCCCTG.......................................................................................................................................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..................................................................................................................................................................................................................................................................................................................................................................................GAGCTTATTCCTGAGGATCTGCA............. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................ATGAAAGGATGAGAGCTGGCTTCTGACA........................................................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................TGAGAGGATGAAAGGATGAGAGCTGG................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................TGAAAGGATGAGAGCTGGCTTCTGAC......................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................................................................................................................................................................AGAAAAGAGAAGACCTGAGCTTATTCC......................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................ACCATGTGACTCACAGCTGAAAGA........................................................................................................................................................................................................................................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...................................................................................................................................................................................................................................................................................................................................................................GAAAAGAGAAGACCTGAGCTTATTCCT........................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......CACCTGAAGGTTCCAGTTCCTGAGGAAA................................................................................................................................................................................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| GTTTCAGCACCTGAAGGTTCCAGTTCCTGAGGAAATCTCCGTGGATCTAGGTAAGTTTGAAGCACCCGAGTCCCTGAAGATTAGTGTCTCCATCAAACTGTGGCTCGCCTCAGATCCGCTGTTTTATGTTAGTTTAGTCAGGCTACTTTAACCATGTGACTCACAGCTGAAAGAGAGTCTACTACACTGAGAGGATGAAAGGATGAGAGCTGGCTTCTGACAGGAAGTGCAGTAACTGCACTGAGTAAGTTCTCTGGTGTAGTAAGTTCAGTGTGGATGTGCAATGTCACTGTGCTGCCAGTGTCAGGCTGCATAGACCATCTCTTTAGATCACTTAATTCCTCTTCTTTGTGCAGAAAAGAGAAGACCTGAGCTTATTCCTGAGGATCTGCATCGCCTCTACATT ................................................................................................................................................................................................................................................................(((....(((...((.((((.((((((.((.(((((......))))).))...))))))..))))))))).....)))........................................................................ ................................................................................................................................................................................................................................................................257................................................................................................356................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | mjTestesWT1() Testes Data. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR014231(GSM319955) 16.5 dpc total. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................................................................................................................................................GTAAGTTCAGTGTGGAccag................................................................................................................................. | 20 | ccag | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................AGCTGGCTTCTGACAGGA..................................................................................................................................................................................... | 18 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - |