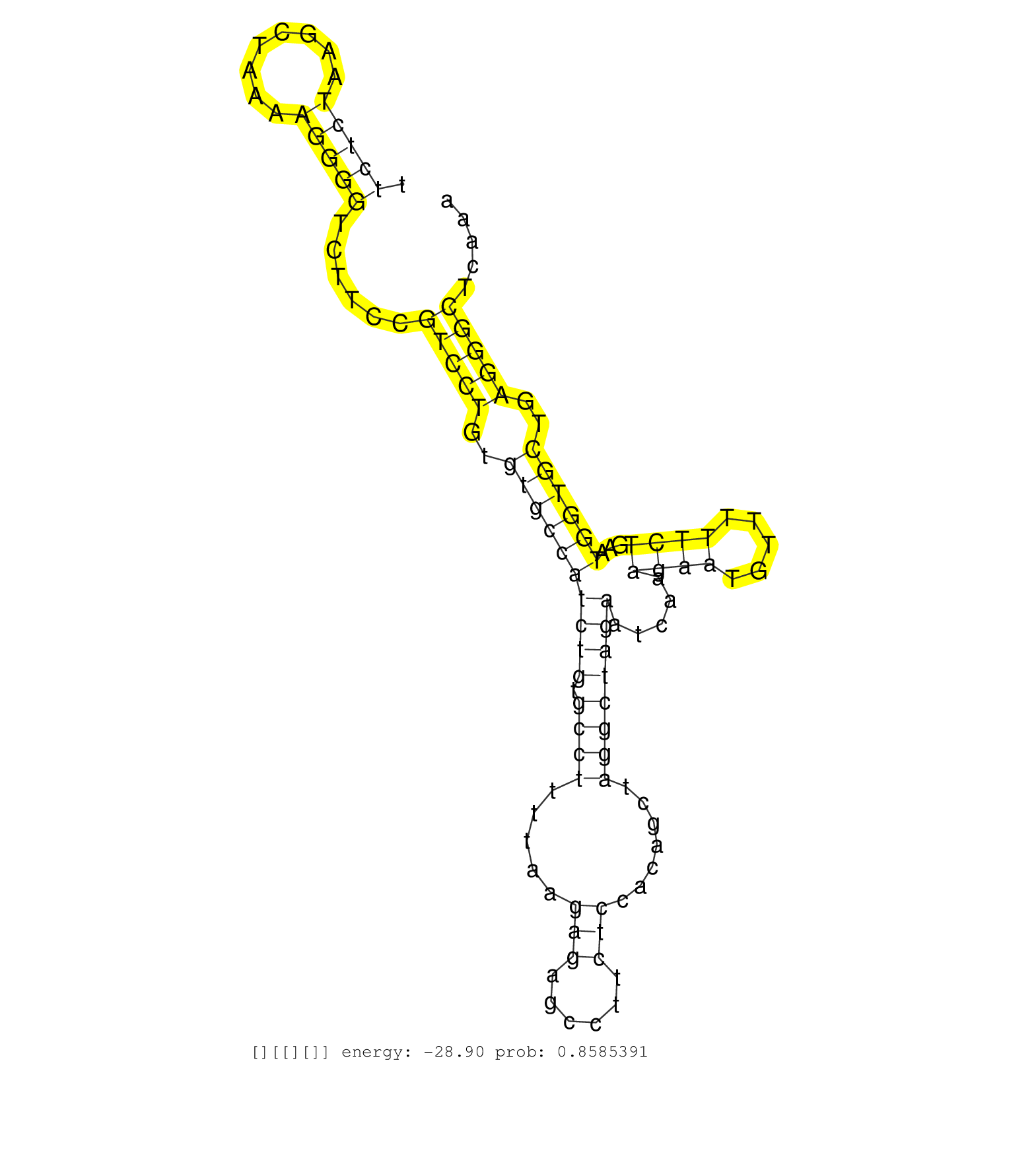
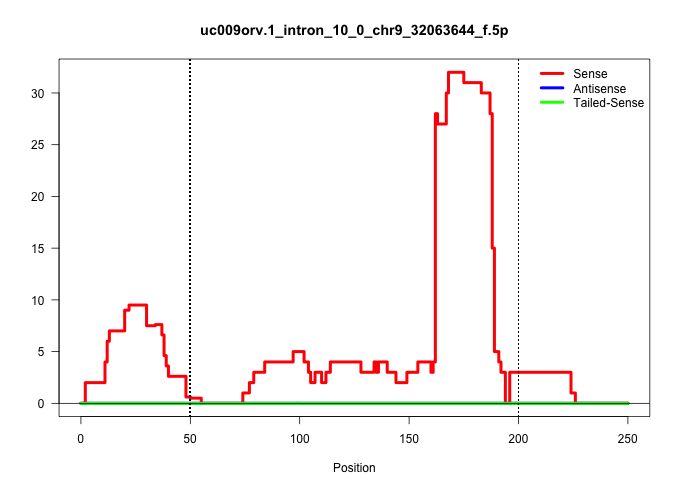
| Gene: Grit | ID: uc009orv.1_intron_10_0_chr9_32063644_f.5p | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(1) OVARY |
(5) PIWI.ip |
(1) PIWI.mut |
(20) TESTES |
| TGTCCTGAGCAGTCAGAGCAAGGCTGTACCGTCTGGACAGAGTCAGACAGGTACTGTTTGTTCTCCTCCGTTCCTTCTCTAAGCTAAAAGGGGTCTTCCGTCCTGTGTGCCATCTGTGCCTTTTAAGAGAGCCTTCTCCACAGCTAGGCTAGAATCAAAGAATGTTTTTCTGAATGGTGCTGAGGGCTCAAAGCCCTAAACTAAAATGAAGCAAACAAAACTCCCCAAAGGGCCCATGGTCCTGCTGCGA ...........................................................................(((((........)))))......(((((..((((((((((.((((.....(((......))).......)))))))).....((((.....))))...))))))..)))))............................................................... ..........................................................................75...................................................................................................................192........................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR014234(GSM319958) Ovary total. (ovary) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................................TGTTTTTCTGAATGGTGCTGAGGGCT.............................................................. | 26 | 1 | 13.00 | 13.00 | 8.00 | 3.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................TGTTTTTCTGAATGGTGCTGAGGGCTC............................................................. | 27 | 1 | 10.00 | 10.00 | 5.00 | 4.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................TCTGGACAGAGTCAGACAGGag..................................................................................................................................................................................................... | 22 | ag | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| ....................AGGCTGTACCGTCTGGACAGAGTCAGAC.......................................................................................................................................................................................................... | 28 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................TTCTGAATGGTGCTGAGGGCTCAAAGC........................................................ | 27 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................TAAACTAAAATGAAGCAAACAAAACTCC.......................... | 28 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..TCCTGAGCAGTCAGAGCAAGGCTGTACC............................................................................................................................................................................................................................ | 28 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................TGTTTTTCTGAATGGTGCTGAGGGC............................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................TCAAAGAATGTTTTTCTGAATGGTGCTGA................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................TAAAAGGGGTCTTCCGTCCTGTGTGC............................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................TAGAATCAAAGAATGTTTTTCTGAAT........................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .............CAGAGCAAGGCTGTACCGTCTGGACA................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................TGCCATCTGTGCCTTTTAAGAGAGCCTT................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................TGAATGGTGCTGAGGGCTCAAAGC........................................................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............TCAGAGCAAGGCTGTACCGTCTGGAC.................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................TCTGAATGGTGCTGAGGGCTCAAAGC........................................................ | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................TGTGCCTTTTAAGAGAGCCTTCTCCACAGC.......................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................TCTGTGCCTTTTAAGAGAGCCTTCTCCA.............................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........GTCAGAGCAAGGCTGTACCGTCTGGAC.................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................TTCTCTAAGCTAAAAGGGGTCTTCCGTC.................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................AGGCTGTACCGTCTGGACAGAGTCAGACAG........................................................................................................................................................................................................ | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ......................................................................................................................................TCTCCACAGCTAGGCTAGAATCAAAG.......................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ............TCAGAGCAAGGCTGTACCGTCTGGACAG.................................................................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................TAAAATGAAGCAAACAAAACTCCCCAAcg.................... | 29 | cg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...............................................................................TAAGCTAAAAGGGGTCTTCCGTCCTG................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................TAAACTAAAATGAAGCAAACAAAACTCCCC........................ | 30 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................TAGGCTAGAATCAAAGAATGTTTTTCTGAA............................................................................ | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .............................................................................TCTAAGCTAAAAGGGGTCTTCCGTCCT.................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................TTCTGAATGGTGCTGAGGGCTCAAA.......................................................... | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................AGGCTGTACCGTCTGG...................................................................................................................................................................................................................... | 16 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................ATGTTTTTCTGAATGGTGCTGAGGGC............................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........GTCAGAGCAAGGCTGTACCGTCTGGA..................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................TCCACAGCTAGGCTAGAATCAAAGAAT....................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................CCGTCCTGTGTGCCATCTGTGCCTTTTAAGA.......................................................................................................................... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ........................................................................................................................................................................TCTGAATGGTGCTGAGGGCTCAA........................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ......................................AGAGTCAGACAGGTACT................................................................................................................................................................................................... | 17 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - |
| ......................GCTGTACCGTCTGGAC.................................................................................................................................................................................................................... | 16 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - |
| ..................................GGACAGAGTCAGACAG........................................................................................................................................................................................................ | 16 | 9 | 0.11 | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.11 |
| TGTCCTGAGCAGTCAGAGCAAGGCTGTACCGTCTGGACAGAGTCAGACAGGTACTGTTTGTTCTCCTCCGTTCCTTCTCTAAGCTAAAAGGGGTCTTCCGTCCTGTGTGCCATCTGTGCCTTTTAAGAGAGCCTTCTCCACAGCTAGGCTAGAATCAAAGAATGTTTTTCTGAATGGTGCTGAGGGCTCAAAGCCCTAAACTAAAATGAAGCAAACAAAACTCCCCAAAGGGCCCATGGTCCTGCTGCGA ...........................................................................(((((........)))))......(((((..((((((((((.((((.....(((......))).......)))))))).....((((.....))))...))))))..)))))............................................................... ..........................................................................75...................................................................................................................192........................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR014234(GSM319958) Ovary total. (ovary) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) |
|---|