
| Gene: Nfrkb | ID: uc009orr.1_intron_11_0_chr9_31208589_f.5p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(4) PIWI.ip |
(1) PIWI.mut |
(12) TESTES |
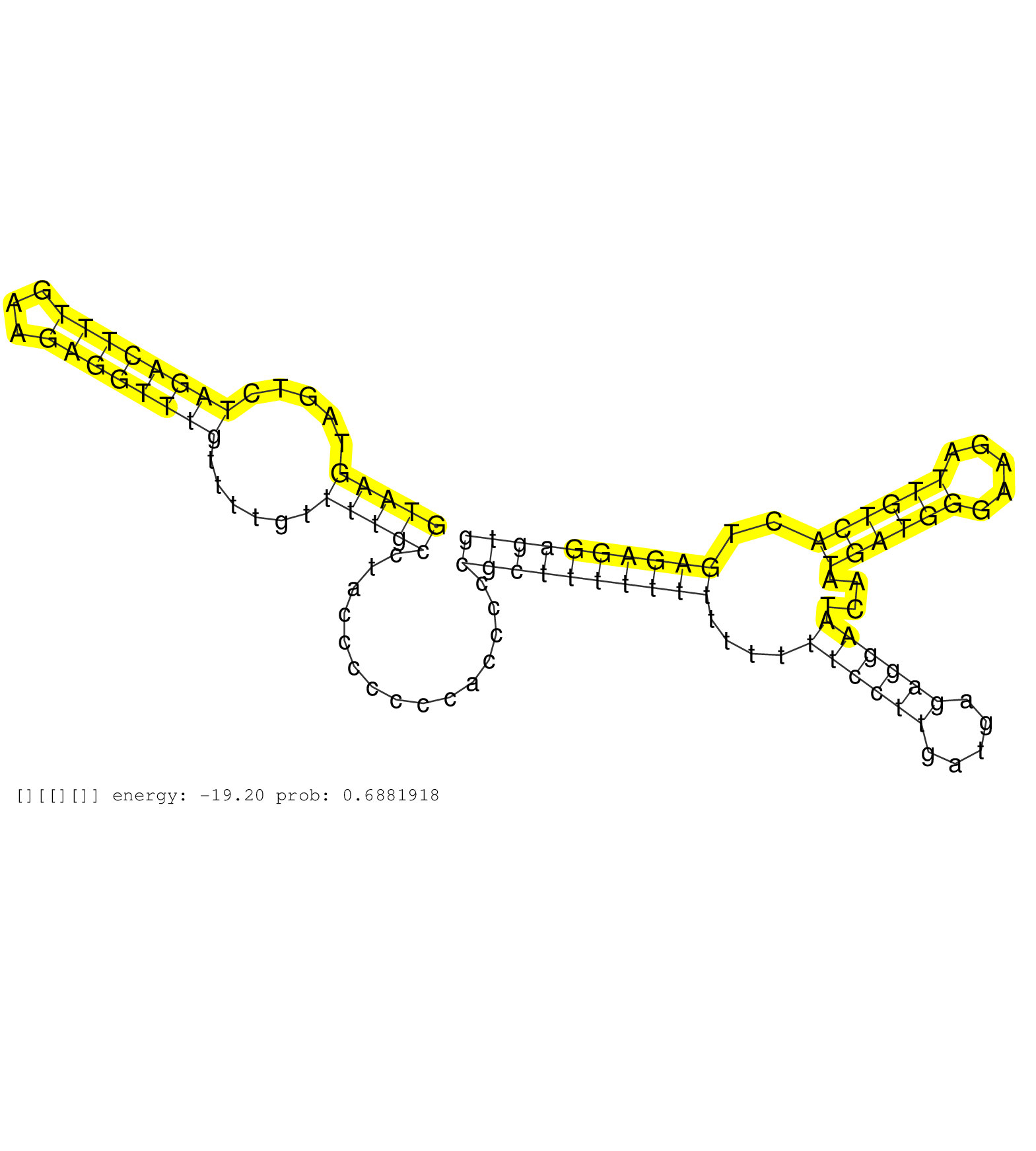
| CTCTCCATTTGTTGAATTCAAAGAGAAAACCCAGCAGTGGAAATTGCTTGGTAAGTAGTCTAGACTTTGAAGAGGTTTGTTTTGTTTTGCCTACCCCCCCACCCCCGCTTTTTTTTTTTTTCCTTGATGAGAGGAATCAATGATGGGAAGATTGTCACTGAGAGGAGTGGCTAGAGAGCCAGGCAAGCATGGAGATGTGCCTTAGCGTTTCTGAGGAGGAACTATATTTCTTGTTAAAGTAGGGGGAAGG ..................................................(((((.....((((((((...))))))))......)))))...............((((((((((....((((((.....))))))....((((((.....))))))..))))))))))................................................................................. ..................................................51....................................................................................................................169............................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | GSM475281(GSM475281) total RNA. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................AATCAATGATGGGAAGATTGTCACTGAGAGG..................................................................................... | 31 | 1 | 6.00 | 6.00 | - | - | 3.00 | - | 1.00 | 1.00 | - | - | - | - | 1.00 | - |
| .......................................................................................................................................ATCAATGATGGGAAGATTGTCACTGAGA....................................................................................... | 28 | 1 | 3.00 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGTAGTCTAGACTTTGAAGAGGTT............................................................................................................................................................................. | 27 | 1 | 3.00 | 3.00 | - | 1.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - |
| .......................................................................................................................................ATCAATGATGGGAAGATTGTCACTGAGAGGA.................................................................................... | 31 | 1 | 3.00 | 3.00 | - | - | 1.00 | - | 1.00 | 1.00 | - | - | - | - | - | - |
| ..................................................GTAAGTAGTCTAGACTTTGAAGAGGT.............................................................................................................................................................................. | 26 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .......................................................................................................................................ATCAATGATGGGAAGATTGTCACTGAGAGG..................................................................................... | 30 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ........TTGTTGAATTCAAAGAGAAAACCCAGCA...................................................................................................................................................................................................................... | 28 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - |
| ......ATTTGTTGAATTCAAAGAGAAAACCCAGt....................................................................................................................................................................................................................... | 29 | t | 2.00 | 0.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - |
| ..................................................GTAAGTAGTCTAGACTTTGAAGAGGTTT............................................................................................................................................................................ | 28 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - |
| ...............ATTCAAAGAGAAAACCCAGCAGTGGAA................................................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ......................................................................................................................................AATCAATGATGGGAAGATTGTCACTGAGAGGA.................................................................................... | 32 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..............................................................................................................................ATGAGAGGAATCAATGATGGGAAGATT................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .........TGTTGAATTCAAAGAGAAAACCCAGCA...................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| CTCTCCATTTGTTGAATTCAAAGAGAAAACCCAGCAGTGGAAATTGCTTGGTAAGTAGTCTAGACTTTGAAGAGGTTTGTTTTGTTTTGCCTACCCCCCCACCCCCGCTTTTTTTTTTTTTCCTTGATGAGAGGAATCAATGATGGGAAGATTGTCACTGAGAGGAGTGGCTAGAGAGCCAGGCAAGCATGGAGATGTGCCTTAGCGTTTCTGAGGAGGAACTATATTTCTTGTTAAAGTAGGGGGAAGG ..................................................(((((.....((((((((...))))))))......)))))...............((((((((((....((((((.....))))))....((((((.....))))))..))))))))))................................................................................. ..................................................51....................................................................................................................169............................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | GSM475281(GSM475281) total RNA. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................................................................................AGCCAGGCAAGCATGagat........................................................... | 19 | agat | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - |