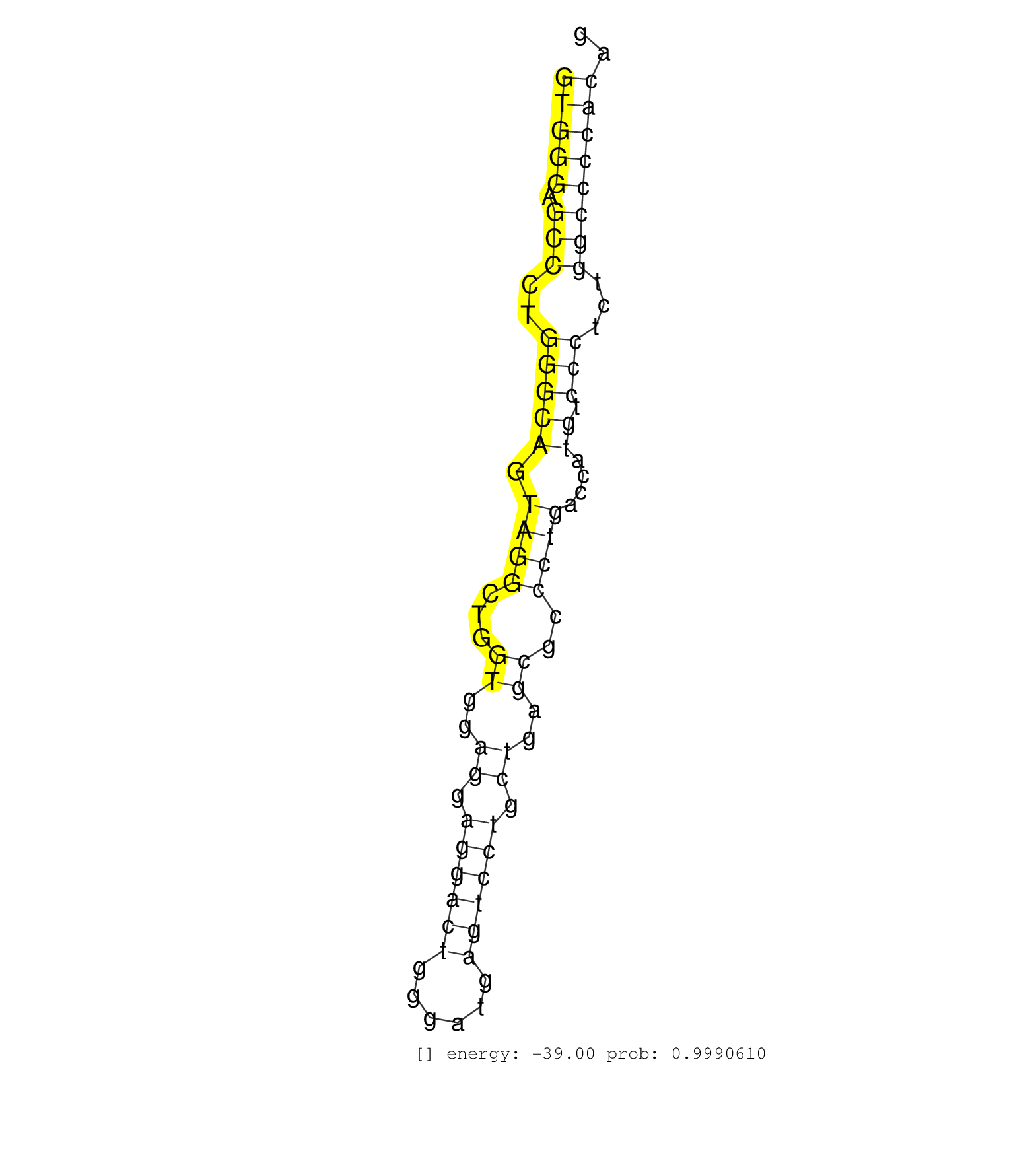

| Gene: Tyk2 | ID: uc009oke.1_intron_16_0_chr9_20924862_r | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(4) PIWI.ip |
(2) PIWI.mut |
(17) TESTES |
| TGCTAAAGGAGCGTCGCGTGCACATCCACCTCCAAGACAACAAGTGCTTGGTGGGAGCCCTGGGCAGTAGGCTGGTGGAGGAGGACTGGGATGAGTCCTGCTGAGCGCCCTGACCATGTCCCTCTGGCCCCACAGTTGCTGTGCCTCTGTTCCCAGGCTGAGGCCCTGTCCTTTGTGGCCCTGGT ..................................................(((((.(((..(((((.((((...((..((.((((((......)))))).))..))..))))....)).)))...)))))))).................................................... ..................................................51..................................................................................135................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTGGGAGCCCTGGGCAGTAGGCTGGT............................................................................................................. | 26 | 1 | 17.00 | 17.00 | 5.00 | 5.00 | 3.00 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - |
| ..................................................GTGGGAGCCCTGGGCAGTAGGCTGG.............................................................................................................. | 25 | 1 | 6.00 | 6.00 | 2.00 | 1.00 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TGGGAGCCCTGGGCAGTAGGCTGGT............................................................................................................. | 25 | 1 | 4.00 | 4.00 | 1.00 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGGGAGCCCTGGGCAGTAGGCTGGa............................................................................................................. | 26 | a | 3.00 | 6.00 | - | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................TGCTGTGCCTCTGTTCCCAGGCTGAGGC..................... | 28 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - |
| ..................................................GTGGGAGCCCTGGGCAGTAGGCTGGTG............................................................................................................ | 27 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...................................................TGGGAGCCCTGGGCAGTAGGCTGGTt............................................................................................................ | 26 | t | 1.00 | 4.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGGGAGCCCTGGGCAGTAGGCTGGTt............................................................................................................ | 27 | t | 1.00 | 17.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGGGAGCCCTGGGCAGTAGG.................................................................................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGGGAGCCCTGGGCAGTAGGCTGGTaga.......................................................................................................... | 29 | aga | 1.00 | 17.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TGGGAGCCCTGGGCAGTAGGCTGGTGGA.......................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGGGAGCCCTGGGCAGTAGGtt................................................................................................................ | 23 | tt | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...TAAAGGAGCGTCGCGTGCACATCCACC........................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..................................................GTGGGAGCCCTGGGCAGTAGGCTGa.............................................................................................................. | 25 | a | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...................................................TGGGAGCCCTGGGCAGTAGGCTGGTGt........................................................................................................... | 27 | t | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGGGAGCCCTGGGCAGTAGGCTGGg............................................................................................................. | 26 | g | 1.00 | 6.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..................................................GTGGGAGCCCTGGGCAGT..................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGGGAGCCCTGGGCAGTAGGCTG............................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................GTGGGAGCCCTGGGCAGTAGGCTaa.............................................................................................................. | 25 | aa | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .....AAGGAGCGTCGCGTGCACATCCACCTC......................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ............................................................TGGGCAGTAGGCTGGTGGAGGAGGACT.................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| TGCTAAAGGAGCGTCGCGTGCACATCCACCTCCAAGACAACAAGTGCTTGGTGGGAGCCCTGGGCAGTAGGCTGGTGGAGGAGGACTGGGATGAGTCCTGCTGAGCGCCCTGACCATGTCCCTCTGGCCCCACAGTTGCTGTGCCTCTGTTCCCAGGCTGAGGCCCTGTCCTTTGTGGCCCTGGT ..................................................(((((.(((..(((((.((((...((..((.((((((......)))))).))..))..))))....)).)))...)))))))).................................................... ..................................................51..................................................................................135................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) |
|---|