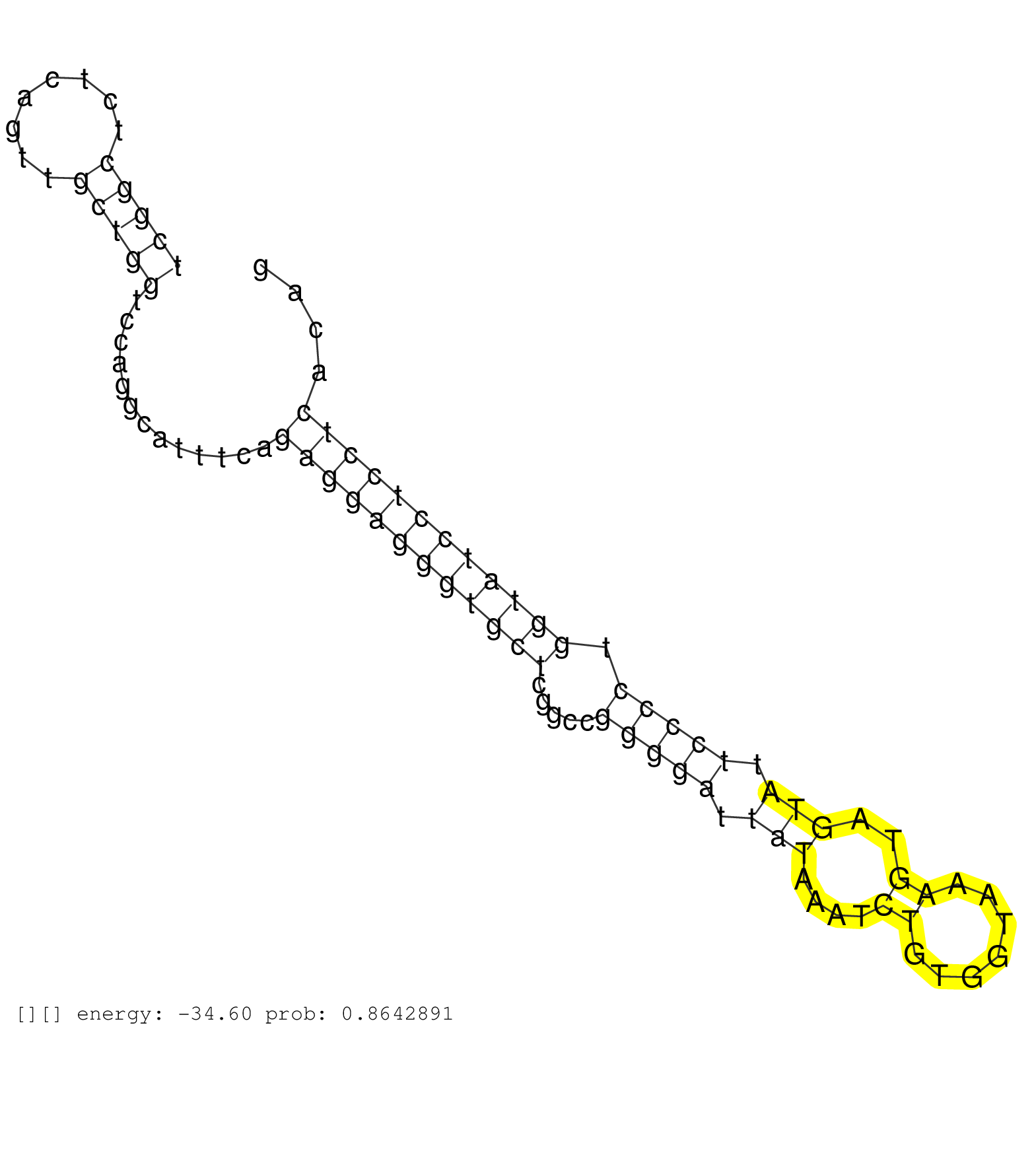

| Gene: Dnmt1 | ID: uc009ojo.1_intron_10_0_chr9_20718534_r.3p | SPECIES: mm9 |
 |
 |
|
(6) OTHER.mut |
(1) PIWI.ip |
(2) PIWI.mut |
(17) TESTES |
| AGGTCGGCTTGGGCAGCAGAGCAAGACACTCTCAACAAATAAAAGGCCTTGAAGCTGTGGTGTGAGGTGTACAAGACTAATAAGATTCCTGTTCTCAGACTCGGCTCTCAGTTGCTGGTCCAGGCATTTCAGAGGAGGGTGCTCGGCCGGGGATTATAAATCTGTGGTAAAGTAGTATTCCCCTGGTATCCTCCTCACAGGCCTGAGAATACCCACAGGTCCTACAACGGATCCTATCACACTGACATCA ....................................................................................................(((((........))))).............((((((((((((.....(((((.(((....((.......))..))).))))).))))))))))))...................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesWT2() Testes Data. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................................................................................................................................TACAACGGATCCTATCACACTGACATC. | 27 | 1 | 8.00 | 8.00 | - | 3.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | 1.00 | - |
| .................................................................................................................................................................................TTCCCCTGGTATCCTCCTCACA................................................... | 22 | 1 | 4.00 | 4.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .................................................................................................................................................................................TTCCCCTGGTATCCTCCgcac.................................................... | 21 | gcac | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................TAAATCTGTGGTAAAGTAGTA......................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...................................................................................................................................................................................................................................CGGATCCTATCACACTGACATC. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................CCTACAACGGATCCTATCACACT....... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TCCCCTGGTATCCTCCTCACtaa................................................. | 23 | taa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................CCCACAGGTCCTACAACGGA................... | 20 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................TCAGAGGAGGGTGCTCGGCCGGGGAT................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................................ACGGATCCTATCACACTGACA... | 21 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................AGAGGAGGGTGCTCGGCCGGGGATT............................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................ACAACGGATCCTATCACACTGACATC. | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................TGAGAATACCCACAGGTCCTACAACGGA................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .........................................................................................................................................GGTGCTCGGCCGGGGATTATAAATCTGTGGTA................................................................................. | 32 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| AGGTCGGCTTGGGCAGCAGAGCAAGACACTCTCAACAAATAAAAGGCCTTGAAGCTGTGGTGTGAGGTGTACAAGACTAATAAGATTCCTGTTCTCAGACTCGGCTCTCAGTTGCTGGTCCAGGCATTTCAGAGGAGGGTGCTCGGCCGGGGATTATAAATCTGTGGTAAAGTAGTATTCCCCTGGTATCCTCCTCACAGGCCTGAGAATACCCACAGGTCCTACAACGGATCCTATCACACTGACATCA ....................................................................................................(((((........))))).............((((((((((((.....(((((.(((....((.......))..))).))))).))))))))))))...................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesWT2() Testes Data. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........AGCAGAGCAAGACACaggc............................................................................................................................................................................................................................. | 19 | aggc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |