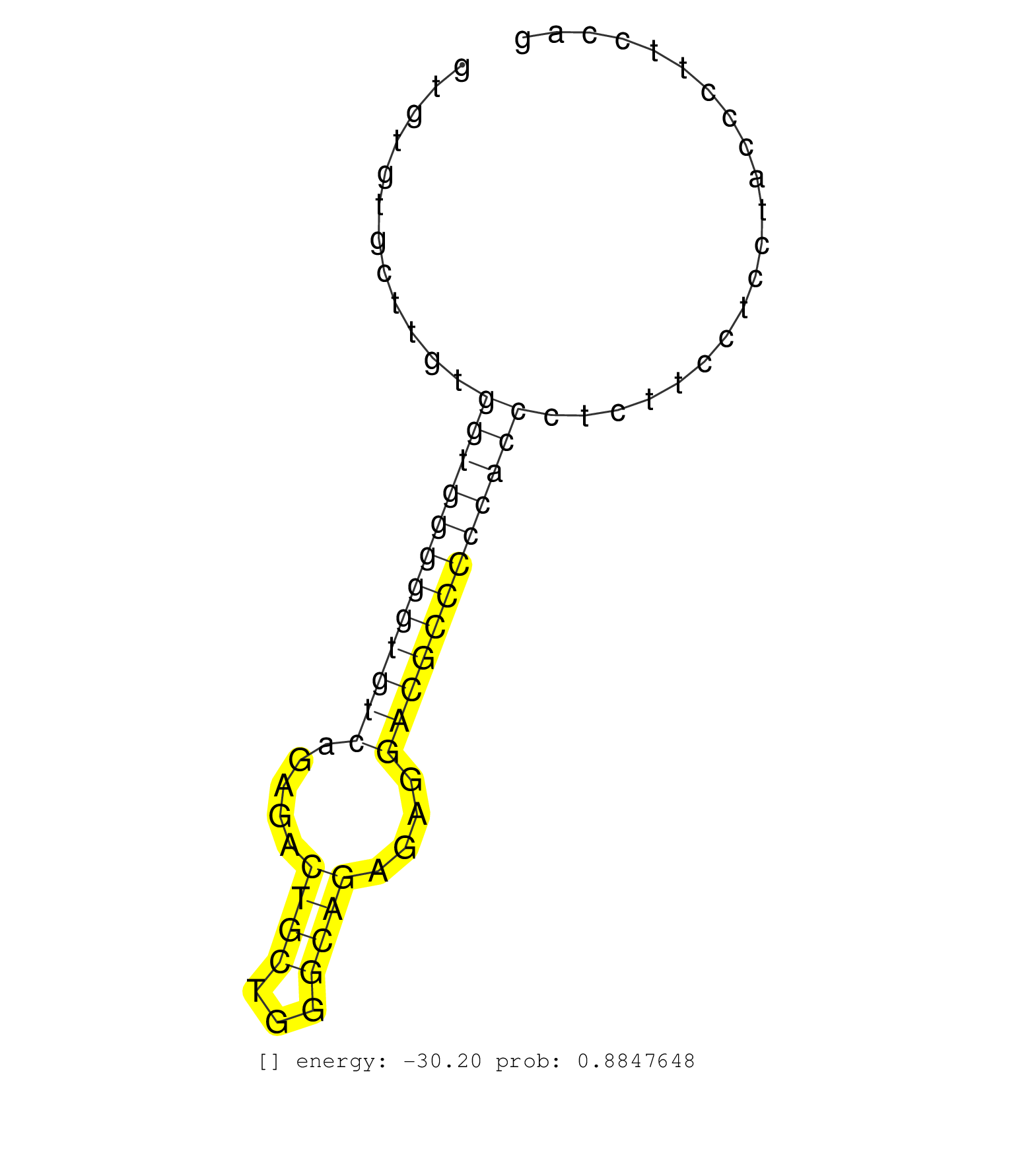
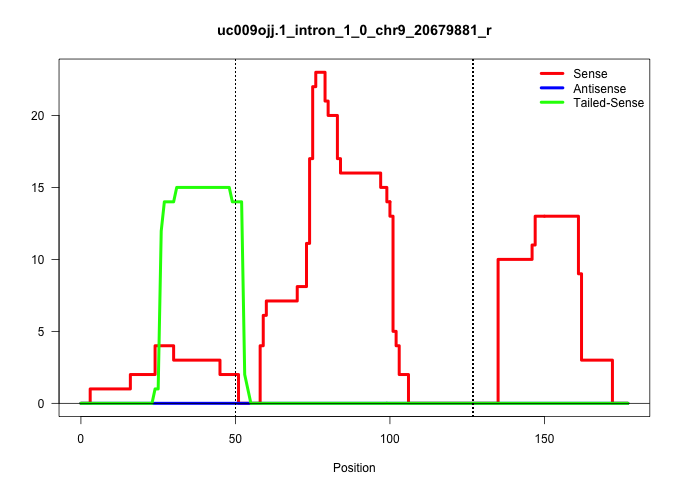
| Gene: Angptl6 | ID: uc009ojj.1_intron_1_0_chr9_20679881_r | SPECIES: mm9 |
 |
 |
|
(1) OTHER.ip |
(5) OTHER.mut |
(5) PIWI.ip |
(3) PIWI.mut |
(24) TESTES |
| GACGGCAGGACGGCTCTGTCAACTTCTTCACCAACTGGCAGCACTACAAGGTGTGTGCTTGTGGTGGGGGTGTCAGAGACTGCTGGGCAGAGAGGACGCCCCCACCCTCTTCCTCCTACCCTTCCAGGCGGGCTTTGGGCGTCCAGAAGGAGAATACTGGCTGGGCCTGGAACCTGT ..............................................................((((((((((((.....((((...))))....))))))))))))....................................................................... ..................................................51..........................................................................127................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesWT4() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | mjTestesWT2() Testes Data. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................TTCACCAACTGGCAGCACTACAAGGcg............................................................................................................................ | 27 | cg | 10.00 | 0.00 | 2.00 | 5.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - |
| ..........................................................TTGTGGTGGGGGTGTCAGAGACTGCT............................................................................................. | 26 | 1 | 8.00 | 8.00 | - | - | 8.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................TGGGCGTCCAGAAGGAGAATACTGGCT............... | 27 | 1 | 6.00 | 6.00 | 1.00 | 3.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................TGGGCGTCCAGAAGGAGAATACTGGC................ | 26 | 1 | 4.00 | 4.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................GAGACTGCTGGGCAGAGAGGACGCCC............................................................................ | 26 | 1 | 4.00 | 4.00 | - | - | - | - | 2.00 | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................GAGACTGCTGGGCAGAGAGGA................................................................................. | 21 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................TTGTGGTGGGGGTGTCAGAGACTGC.............................................................................................. | 25 | 1 | 3.00 | 3.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...................................................................................................................................................AGGAGAATACTGGCTGGGCCTGGAA..... | 25 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................TCTTCACCAACTGGCAGCACTACAAGG.............................................................................................................................. | 27 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................TGTGGTGGGGGTGTCAGAGA.................................................................................................. | 20 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................AGAGACTGCTGGGCAGAGAGGACGCCC............................................................................ | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .........................................................................CAGAGACTGCTGGGCAGAGAGGACGCCC............................................................................ | 28 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................AGAGACTGCTGGGCAGAGAGGACGCCCCCACC....................................................................... | 32 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................TTGTGGTGGGGGTGTCAGAGAC................................................................................................. | 22 | 1 | 2.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...........................TCACCAACTGGCAGCACTACAAGGcgt........................................................................................................................... | 27 | cgt | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................CAACTGGCAGCACTgtgt................................................................................................................................ | 18 | gtgt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................TGTCAGAGACTGCTGGGCAGAGAGGAC................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................AGAGACTGCTGGGCAGAGAGGACGCCCCC.......................................................................... | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................TTCACCAACTGGCAGCACTACAAGGcggg.......................................................................................................................... | 29 | cggg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................TCACCAACTGGCAGCACTACAAGGcg............................................................................................................................ | 26 | cg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ............................................................................AGACTGCTGGGCAGAGAGGACGCCCCC.......................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................CAGAGACTGCTGGGCAGAGAGGACGC.............................................................................. | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................GAGACTGCTGGGCAGAGAGGACGCCCC........................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................TGTGGTGGGGGTGTCAGAGAC................................................................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..................................................................................................................................................AAGGAGAATACTGGCTGGGCCTGGAA..... | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................AGAGACTGCTGGGCAGAGAGGACGCCCCCA......................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...GGCAGGACGGCTCTGTCAACTTCTTCA................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ............................................................GTGGTGGGGGTGTCAGAGACTGCT............................................................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ........................TCTTCACCAACTGGCAGCACTACAAGGcg............................................................................................................................ | 29 | cg | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................TGTCAACTTCTTCACCAACTGGCAGCACT.................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................AGAGACTGCTGGGCAGAGAGGACGCC............................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................TGTGGTGGGGGTGTC....................................................................................................... | 15 | 9 | 0.11 | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.11 |
| GACGGCAGGACGGCTCTGTCAACTTCTTCACCAACTGGCAGCACTACAAGGTGTGTGCTTGTGGTGGGGGTGTCAGAGACTGCTGGGCAGAGAGGACGCCCCCACCCTCTTCCTCCTACCCTTCCAGGCGGGCTTTGGGCGTCCAGAAGGAGAATACTGGCTGGGCCTGGAACCTGT ..............................................................((((((((((((.....((((...))))....))))))))))))....................................................................... ..................................................51..........................................................................127................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesWT4() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | mjTestesWT2() Testes Data. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) |
|---|