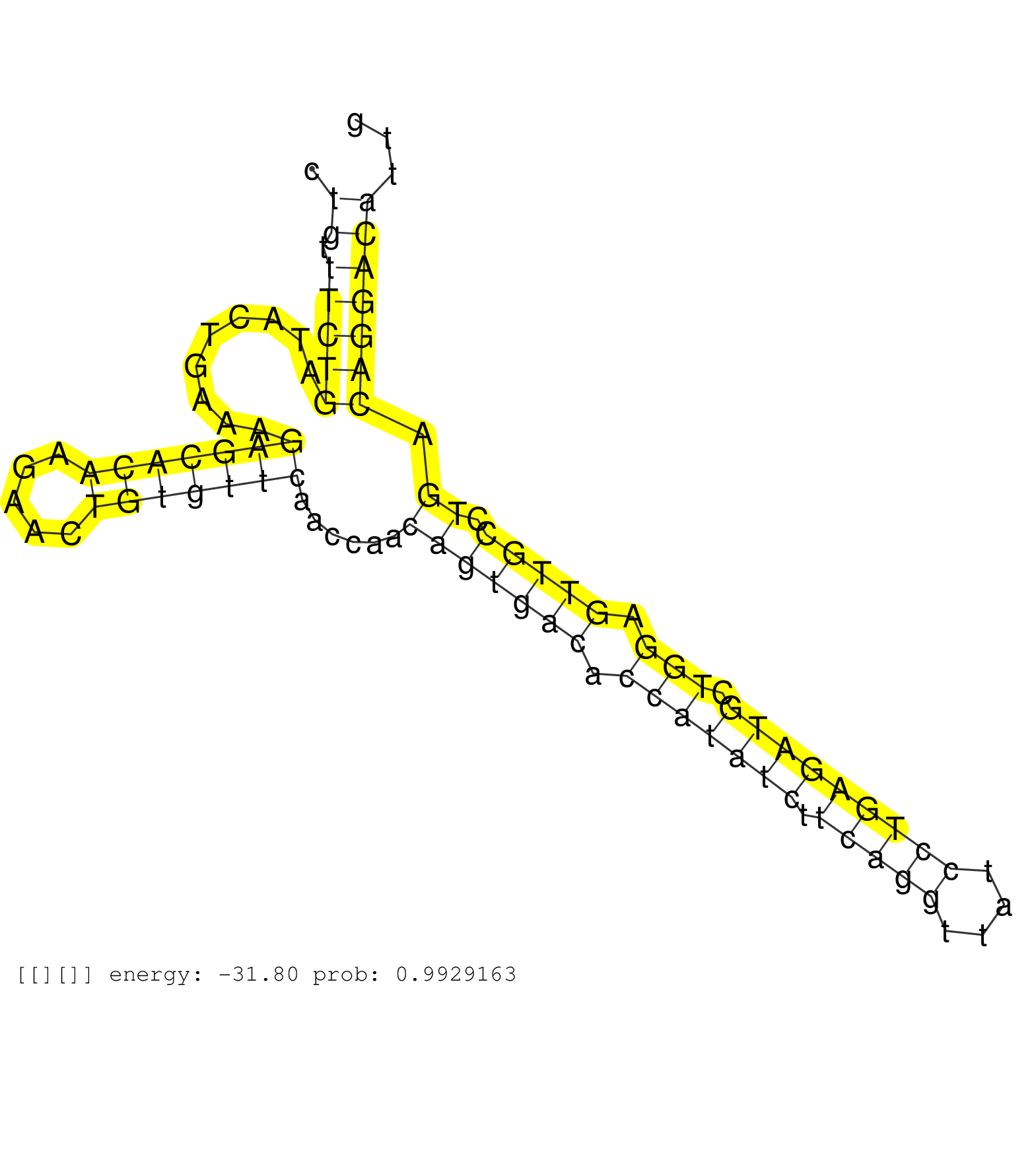

| Gene: 5830418K08Rik | ID: uc009ofv.1_intron_11_0_chr9_15135355_r.5p | SPECIES: mm9 |
 |
 |
|
(4) OTHER.mut |
(4) PIWI.ip |
(1) PIWI.mut |
(14) TESTES |
| CTCATGAAGAAACACTAAGAGAATGGAAGGAAAAAATATTCCCAGAGCAGGTTGGTCCATTTTCTCCCCTGATACCACAACATTCACTTGCTTCATTCCCTGTTTCTGATACTGAAAGAGCACAAGAACTGTGTTCAACCAACAGTGACACCATATCTTCAGGTTATCCTGAGATGCTGGAGTTGCCTGACAGGACATTGGGTTTATCACATACTGCTTTACCTCAGCAGAATAATCTGACTGCACATCC ....................................................................................................((.(((((.........(((((((.....)))))))......(((((((.(((((((.(((((....))))))))).))).))))).)).)))))))..................................................... ...................................................................................................100..................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................................................................TGAGATGCTGGAGTTGCCTGACAGGAC...................................................... | 27 | 1 | 6.00 | 6.00 | 2.00 | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................TCTGATACTGAAAGAGCACAAGAACTG....................................................................................................................... | 27 | 1 | 5.00 | 5.00 | - | 1.00 | 2.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - |
| ........................................................................................................TCTGATACTGAAAGAGCACAAGAACTGT...................................................................................................................... | 28 | 1 | 4.00 | 4.00 | 1.00 | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................TTCTGATACTGAAAGAGCACAAGAACTGT...................................................................................................................... | 29 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................TTCTGATACTGAAAGAGCACAAGAACTG....................................................................................................................... | 28 | 1 | 2.00 | 2.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .TCATGAAGAAACACTAAGAGAATGGAAGG............................................................................................................................................................................................................................ | 29 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................CAGGACATTGGGTTTgaa.......................................... | 18 | gaa | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .......................................................................................TTGCTTCATTCCCTGTTTCTGATACTGAAA..................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .................................................................................ATTCACTTGCTTCATTCC....................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ...............................................................................................................................................................................................AGGACATTGGGTTTgac.......................................... | 17 | gac | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................TCAGGTTATCCTGAGATGCTGGAGTTGC................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................GCTGGAGTTGCCTGACAGGACATTGGt................................................ | 27 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ......................................................................................................................................................................TCCTGAGATGCTGGAGTTGCCTGACA.......................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .......................................TCCCAGAGCAGGTTGGTCCATTTTCTCC....................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................TGGAAGGAAAAAATATTCCCAGAGCAGGg...................................................................................................................................................................................................... | 29 | g | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................AGATGCTGGAGTTGgtat............................................................. | 18 | gtat | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..........................................................................................................................CAAGAACTGTGTTCAACCAACAGTGACAC................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................TGATACTGAAAGAGCACAAGAACT........................................................................................................................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ........................................................................TACCACAACATTCACTTGatct............................................................................................................................................................ | 22 | atct | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...........................................................................................................................................CAACAGTGACACCATATCTTCAGGTTATCC................................................................................. | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ....TGAAGAAACACTAAGAGAATGGAAGGAA.......................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................TTCCCAGAGCAGGTTGGTCCATTTT........................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ......................................................................................................................................................................TCCTGAGATGCTGGAGTTGCCTGAC........................................................... | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................TTCATTCCCTGTTTCTGATACTGAAA..................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................AGGACATTGGGTTTgaaa......................................... | 18 | gaaa | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| CTCATGAAGAAACACTAAGAGAATGGAAGGAAAAAATATTCCCAGAGCAGGTTGGTCCATTTTCTCCCCTGATACCACAACATTCACTTGCTTCATTCCCTGTTTCTGATACTGAAAGAGCACAAGAACTGTGTTCAACCAACAGTGACACCATATCTTCAGGTTATCCTGAGATGCTGGAGTTGCCTGACAGGACATTGGGTTTATCACATACTGCTTTACCTCAGCAGAATAATCTGACTGCACATCC ....................................................................................................((.(((((.........(((((((.....)))))))......(((((((.(((((((.(((((....))))))))).))).))))).)).)))))))..................................................... ...................................................................................................100..................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) |
|---|