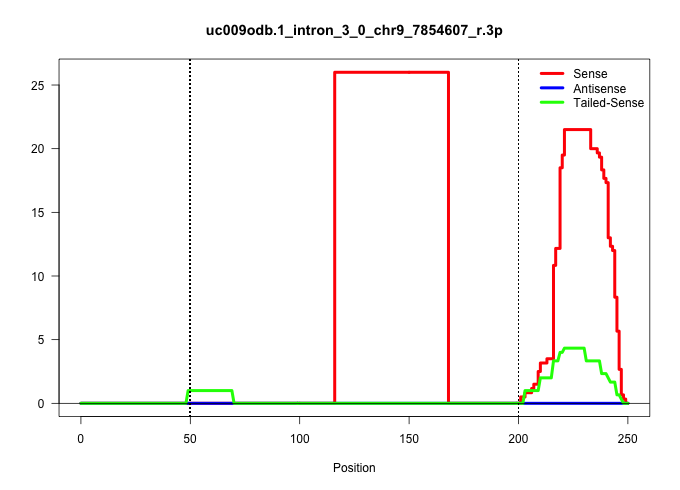
| Gene: Birc3 | ID: uc009odb.1_intron_3_0_chr9_7854607_r.3p | SPECIES: mm9 |
 |
|
(1) OTHER.ip |
(1) OTHER.mut |
(5) PIWI.ip |
(2) PIWI.mut |
(1) TDRD1.ip |
(25) TESTES |
| TTTAAAAAGTATATGAAGCCAGATATGTTGGTTTGTGCCTAGCACTTTCTGAGGCAGGAGAATTGGCATGAATTTGAGATCAGCTTGGAATATATAATAAGCTTGAGGCTAGACTGGCCTAGAAGGATTCCTAAAAGAACAACCACAACAAAAAGATGCATGGGCGCTGCTAATATCTGTTAAATCCCCTGTCCATATAGTCGTGCATTTTGGCCCTGGAGAAAGTTCGGAAGATGTCGTCATGATGAGC |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR363960(GSM822762) Adult Library#1Small RNA Miwi IPDuplexed run:. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................GCCTAGAAGGATTCCTAAAAGAACAACCACAACAAAAAGATGCATGGGCGCT.................................................................................. | 52 | 1 | 26.00 | 26.00 | 26.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................TGGAGAAAGTTCGGAAGATGTCGTC......... | 25 | 3 | 4.33 | 4.33 | - | 1.00 | 1.00 | 0.33 | 0.33 | 0.67 | 0.33 | - | - | - | - | - | 0.33 | - | - | - | - | 0.33 | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................AGAAAGTTCGGAAGATGTCGTCATGAT.... | 27 | 3 | 2.33 | 2.33 | - | - | 1.00 | - | - | - | 1.00 | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................AGAAAGTTCGGAAGATGTCGTCATGA..... | 26 | 3 | 2.33 | 2.33 | - | 0.33 | 1.00 | - | 0.67 | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................TGGAGAAAGTTCGGAAGATGTCGTCATG...... | 28 | 3 | 1.67 | 1.67 | - | 0.33 | 0.33 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................AAAGTTCGGAAGATGTCGTCATGATG... | 26 | 3 | 1.67 | 1.67 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | 0.33 | - | - | - | - | - |
| .................................................TGAGGCAGGAGAATTGGgatt.................................................................................................................................................................................... | 21 | gatt | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................TGCATTTTGGCCCTGGAGAAAGTTCGGt................... | 28 | t | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................GGAGAAAGTTCGGAAGATGTCGTCATG...... | 27 | 3 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 0.33 | - | - | 0.33 | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - |
| ...........................................................................................................................................................................................................................AGAAAGTTCGGAAGATGTCGTCATG...... | 25 | 3 | 1.00 | 1.00 | - | 0.33 | - | - | - | - | - | - | - | - | - | 0.33 | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................TGGCCCTGGAGAAAGTTCGGAAGATGTa............ | 28 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................TGGCCCTGGAGAAAGTTCGGAAGATGTC............ | 28 | 3 | 0.67 | 0.67 | - | 0.33 | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................TGGAGAAAGTTCGGAAGATGTCGTCATGt..... | 29 | t | 0.67 | 1.67 | - | - | - | 0.33 | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................GAAAGTTCGGAAGATGTCGTCATGAT.... | 26 | 3 | 0.67 | 0.67 | - | 0.33 | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................CGTGCATTTTGGCCCTGGAGAAAGTTCGGAAG................. | 32 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................TTGGCCCTGGAGAAAGTTCGGAAGATGTC............ | 29 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................AAAGTTCGGAAGATGTCGTCATGATGAG. | 28 | 3 | 0.33 | 0.33 | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................TGGAGAAAGTTCGGAAGATGTCG........... | 23 | 3 | 0.33 | 0.33 | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................AGAAAGTTCGGAAGATGTCGTCATGATt... | 28 | t | 0.33 | 2.33 | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................TGGAGAAAGTTCGGAAGATGTCGT.......... | 24 | 3 | 0.33 | 0.33 | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................TTGGCCCTGGAGAAAGTTCGGAAGATGT............. | 28 | 3 | 0.33 | 0.33 | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................ATTTTGGCCCTGGAGAAAGTTCGGAAG................. | 27 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................TGCATTTTGGCCCTGGAGAAAGTTCGGAAG................. | 30 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................AAAGTTCGGAAGATGTCGTCATGATGt.. | 27 | t | 0.33 | 1.67 | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................TTTTGGCCCTGGAGAAAGTTCGGAAG................. | 26 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - |
| .....................................................................................................................................................................................................................CCCTGGAGAAAGTTCGGAAGATGTCG........... | 26 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - |
| ........................................................................................................................................................................................................................TGGAGAAAGTTCGGAAGATGTCGcc......... | 25 | cc | 0.33 | 0.33 | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................AGAAAGTTCGGAAGATGTCGTCATGATGA.. | 29 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................TGGAGAAAGTTCGGAAGATGTCGTCt........ | 26 | t | 0.33 | 4.33 | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................AGAAAGTTCGGAAGATGTCGTCA........ | 23 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................GAAAGTTCGGAAGATGTCGTCATGATG... | 27 | 3 | 0.33 | 0.33 | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................TGGAGAAAGTTCGGAAGATGTCGTCATGA..... | 29 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - |
| ........................................................................................................................................................................................................................TGGAGAAAGTTCGGAAGATGTCGTCA........ | 26 | 3 | 0.33 | 0.33 | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................AGAAAGTTCGGAAGATGTCGTCATGt..... | 26 | t | 0.33 | 1.00 | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................GGAGAAAGTTCGGAAGATGTCGTCAT....... | 26 | 3 | 0.33 | 0.33 | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................TTGGCCCTGGAGAAAGTTCGGAAGATG.............. | 27 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 |
| TTTAAAAAGTATATGAAGCCAGATATGTTGGTTTGTGCCTAGCACTTTCTGAGGCAGGAGAATTGGCATGAATTTGAGATCAGCTTGGAATATATAATAAGCTTGAGGCTAGACTGGCCTAGAAGGATTCCTAAAAGAACAACCACAACAAAAAGATGCATGGGCGCTGCTAATATCTGTTAAATCCCCTGTCCATATAGTCGTGCATTTTGGCCCTGGAGAAAGTTCGGAAGATGTCGTCATGATGAGC |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR363960(GSM822762) Adult Library#1Small RNA Miwi IPDuplexed run:. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................GGTTTGTGCCTAGCccct............................................................................................................................................................................................................... | 18 | ccct | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |