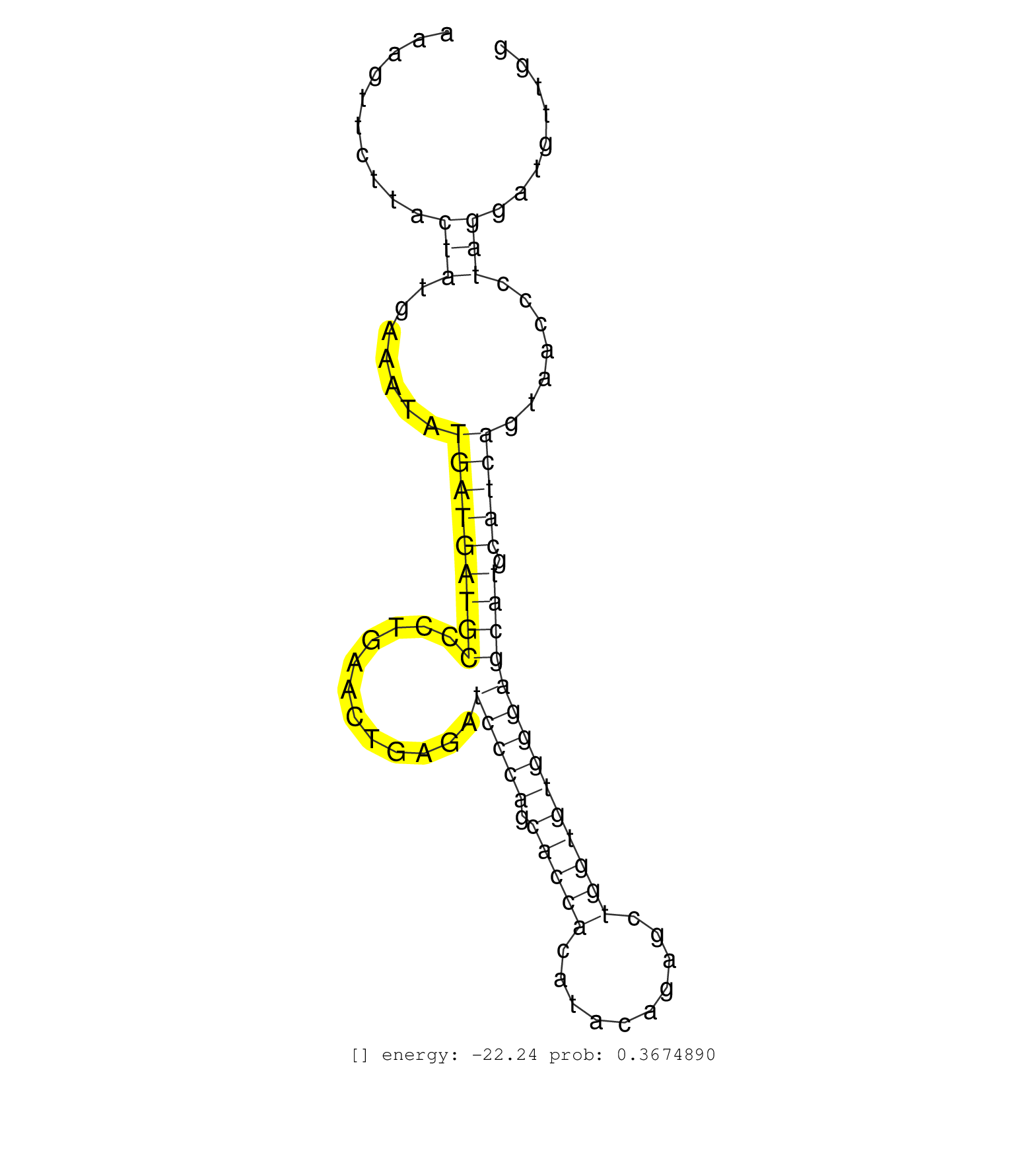

| Gene: Birc2 | ID: uc009ocz.1_intron_4_0_chr9_7825664_r.5p | SPECIES: mm9 |
 |
 |
|
(5) OTHER.mut |
(2) PIWI.ip |
(18) TESTES |
| AGGAGTTTGTTGATGAGATTCAAGCTAGATATCCTCATCTTCTTGAGCAGGTAAACTGATTTTAACATTTACGTGAAAGCGGGGAGATGGCTCATTTGATAAAGTTCTTACTATGAAATATGATGATGCCCTGAACTGAGATCCCAGCACCACATACAGAGCTGGTGTGGGAGCATGCATCAGTAACCCTAGGATGTTGGAAACAGGTGACCCCTGGAGGTCACTGGCCAGGCCATTCAGTGAGTTCCAG ..............................................................................................................(((.......(((((((((............(((((.(((((..........)))))))))))))).))))).......))).......................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT1() Testes Data. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | GSM475281(GSM475281) total RNA. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | mjTestesWT4() Testes Data. (testes) | mjTestesWT2() Testes Data. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................AAATATGATGATGCCCTGAACTGAGA............................................................................................................. | 26 | 1 | 41.00 | 41.00 | 15.00 | 11.00 | 5.00 | 4.00 | 1.00 | - | 1.00 | 1.00 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - |
| ..................................................................................................................GAAATATGATGATGCCCTGAACTGAGA............................................................................................................. | 27 | 1 | 23.00 | 23.00 | 3.00 | 2.00 | 4.00 | - | 5.00 | 4.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | 1.00 | - | 1.00 |
| ...................................................................................................................AAATATGATGATGCCCTGAAC.................................................................................................................. | 21 | 1 | 11.00 | 11.00 | 4.00 | 2.00 | 1.00 | 3.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..................................................................................................................GAAATATGATGATGCCCTGAAC.................................................................................................................. | 22 | 1 | 2.00 | 2.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................AAATATGATGATGCCCTGAA................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................AAATATGATGATGCCCTGAACTGAGAT............................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .........................TAGATATCCTCATCTTCTTGAGCAGct...................................................................................................................................................................................................... | 27 | ct | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .........................TAGATATCCTCATCTTCTTGAGCAGctgt.................................................................................................................................................................................................... | 29 | ctgt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ......................AGCTAGATATCCTCATCTTCTTGAGCAG........................................................................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..................................................................................................................GAAATATGATGATGCCCTGAACTGAGAa............................................................................................................ | 28 | a | 1.00 | 23.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................TATGATGATGCCCTGAACTGAGA............................................................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................AATATGATGATGCCCTGAACT................................................................................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ....................CAAGCTAGATATCCTCATCTTCTTGAGC.......................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..............GAGATTCAAGCTAGATATCCT....................................................................................................................................................................................................................... | 21 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - |
| ..........TGATGAGATTCAAGCTAGATA........................................................................................................................................................................................................................... | 21 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - |
| AGGAGTTTGTTGATGAGATTCAAGCTAGATATCCTCATCTTCTTGAGCAGGTAAACTGATTTTAACATTTACGTGAAAGCGGGGAGATGGCTCATTTGATAAAGTTCTTACTATGAAATATGATGATGCCCTGAACTGAGATCCCAGCACCACATACAGAGCTGGTGTGGGAGCATGCATCAGTAACCCTAGGATGTTGGAAACAGGTGACCCCTGGAGGTCACTGGCCAGGCCATTCAGTGAGTTCCAG ..............................................................................................................(((.......(((((((((............(((((.(((((..........)))))))))))))).))))).......))).......................................................... ....................................................................................................101................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT1() Testes Data. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | GSM475281(GSM475281) total RNA. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | mjTestesWT4() Testes Data. (testes) | mjTestesWT2() Testes Data. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|