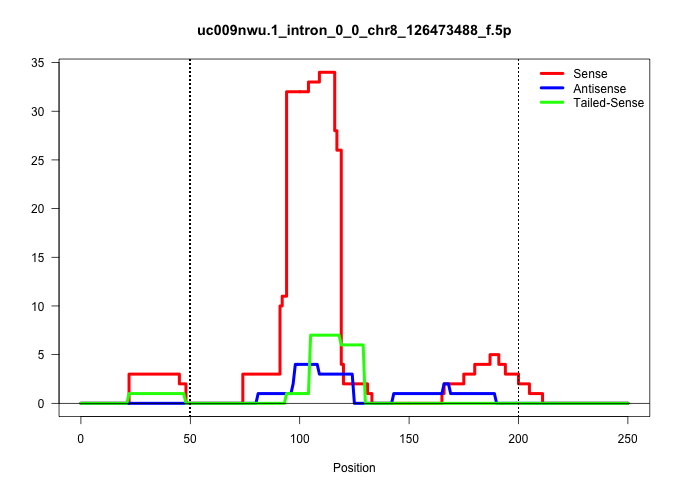
| Gene: AK013187 | ID: uc009nwu.1_intron_0_0_chr8_126473488_f.5p | SPECIES: mm9 |
 |
|
(2) OTHER.mut |
(4) PIWI.ip |
(3) PIWI.mut |
(1) TDRD1.ip |
(20) TESTES |
| AGGAGGCGCCCGTGGGCGGAGCTGAGGAAGTCGCGCTCTGGGCGGCGGTGGTGTGCTCACAGTTTTTCCCAACCTAATTCAGATCGGTTCAGGCTTGTTCTTCGTTCGATTGTCTGGGCTCTGCAGCGGGTTGCAGCTTTTTGCTTTCCAGGCACAGCTTGCATTTTGATGGGTCTGTCCTACTGCCGGAGATTCGGAAATGACAGTGCCCCCAGTCCCCGGGCATGTTCACTCTCATCGCTGCCCCTGA |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT2() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT1() Testes Data. (testes) | mjTestesWT4() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................TTGTTCTTCGTTCGATTGTCTGGGC................................................................................................................................... | 25 | 1 | 21.00 | 21.00 | 8.00 | 6.00 | - | - | 5.00 | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...........................................................................................GGCTTGTTCTTCGTTCGATTGTCTG...................................................................................................................................... | 25 | 1 | 6.00 | 6.00 | - | - | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................TCGATTGTCTGGGCTCTGCAGCGGa........................................................................................................................ | 25 | a | 6.00 | 0.00 | - | - | - | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................TAATTCAGATCGGTTCAGGC............................................................................................................................................................ | 20 | 1 | 3.00 | 3.00 | - | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................TTGTTCTTCGTTCGATTGTCTGGGCT.................................................................................................................................. | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ......................TGAGGAAGTCGCGCTCTGGGCGGCGG.......................................................................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TACTGCCGGAGATTCGGAAATGACA............................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ........................................................................................................TTCGATTGTCTGGGCTCTGCAGCGGGT....................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...........................................................................................................................................................................................GGAGATTCGGAAATGACAGTGCCC....................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...........................................................................................GGCTTGTTCTTCGTTCGATTGTCTGG..................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................TTGTTCTTCGTTCGATTGTCTGGGt................................................................................................................................... | 25 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ............................................................................................GCTTGTTCTTCGTTCGATTGTCTGGGC................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................TGAGGAAGTCGCGCTCTGGGCGGCGt.......................................................................................................................................................................................................... | 26 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................TTGTTCTTCGTTCGATTGTCTGG..................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................TGATGGGTCTGTCCTACTGCCGGAGATT........................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...............................................................................................................................................................................TGTCCTACTGCCGGAGATTCGGAAA.................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .....................................................................................................................................................................TTGATGGGTCTGTCCTACTGCCGGAG........................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................TGAGGAAGTCGCGCTCTGGGCGG............................................................................................................................................................................................................. | 23 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................TTGTCTGGGCTCTGCAGCGGGTTG..................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| AGGAGGCGCCCGTGGGCGGAGCTGAGGAAGTCGCGCTCTGGGCGGCGGTGGTGTGCTCACAGTTTTTCCCAACCTAATTCAGATCGGTTCAGGCTTGTTCTTCGTTCGATTGTCTGGGCTCTGCAGCGGGTTGCAGCTTTTTGCTTTCCAGGCACAGCTTGCATTTTGATGGGTCTGTCCTACTGCCGGAGATTCGGAAATGACAGTGCCCCCAGTCCCCGGGCATGTTCACTCTCATCGCTGCCCCTGA |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT2() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT1() Testes Data. (testes) | mjTestesWT4() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................TCTTCGTTCGATTGTCTGGGCTCTGCA............................................................................................................................. | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - |
| ......................................................................................................................................................................TGATGGGTCTGTCCTACTGCCGGA............................................................ | 24 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................CTTTCCAGGCACAGCTTGCATTTTGA................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .................................................................................GATCGGTTCAGGCTTGTTCTTCGTTCGA............................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................TTCTTCGTTCGATTGTCTGGGCTCTGCA............................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |