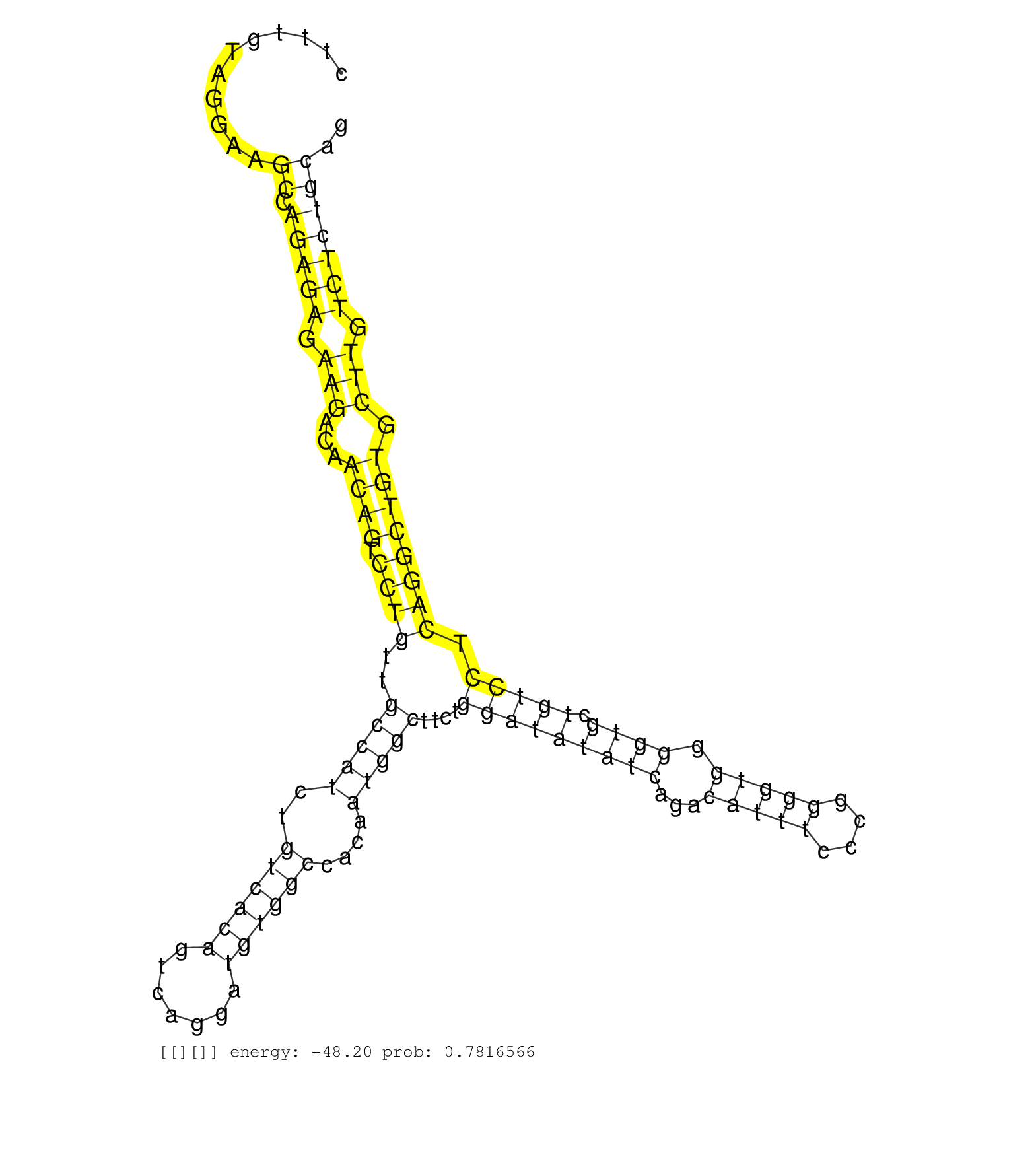
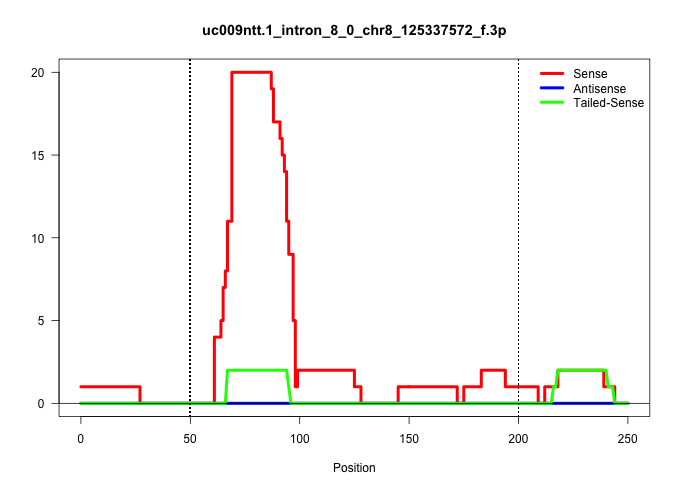
| Gene: BC021611 | ID: uc009ntt.1_intron_8_0_chr8_125337572_f.3p | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(4) PIWI.ip |
(14) TESTES |
| TCTGGATGAGGAGAGCAAAGGACTCGTCCCCTCCTCTCAGTGTCCTCACCCATGTCACTGCTGACTTTGTAGGAAGCCAGAGAGAAGACAACAGTCCTGTTGCCATCTGTCACAGTCAGGATGTGGCCACAATGGCTTCTGGATATATCAGACATTTCCCGGGGTGGGGTGCTGTCCTCAGGCTGTGCTTGTCTCTGCAGAGGTGTCCTGGCCCCCTATGCTGTGCCCTCGGAGCTCCTGCTGGTGGAGG ...........................................................................((.(((((.(((...((((.((((..(((((..((((((.......))))))....)))))....(((((((((...(((((....))))).)))).))))).)))))))).))).))))))).................................................... ................................................................65.....................................................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT4() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................TAGGAAGCCAGAGAGAAGACAACAGTCCT........................................................................................................................................................ | 29 | 1 | 4.00 | 4.00 | 1.00 | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - |
| .....................................................................TAGGAAGCCAGAGAGAAGACAACAGTCC......................................................................................................................................................... | 28 | 1 | 4.00 | 4.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................TGACTTTGTAGGAAGCCAGAGAGAAGA.................................................................................................................................................................. | 27 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................TGTAGGAAGCCAGAGAGAAGACAACAGT........................................................................................................................................................... | 28 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................TGCCATCTGTCACAGTCAGGATGTGGCC.......................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .......................................................................................................................................................................................TGTGCTTGTCTCTGCAGAGGTGTCCT......................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................TTTGTAGGAAGCCAGAGAGAAGACAA............................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................TGCTGTGCCCTCGGAGCTCCTGCTGt...... | 26 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ........................................................................................................................................................................................................................TATGCTGTGCCCTCGGAGCTCCTGt......... | 25 | t | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................TGTAGGAAGCCAGAGAGAAGACAACAG............................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................TGCTGTGCCCTCGGAGCTCCTGCTGG...... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .................................................................TTTGTAGGAAGCCAGAGAGAAGACAACA............................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................CCCCTATGCTGTGCCCTCGGAGCTCCT........... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................TGTAGGAAGCCAGAGAGAAGACAAaagt........................................................................................................................................................... | 28 | aagt | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................TAGGAAGCCAGAGAGAAGACAACAGTCCTGT...................................................................................................................................................... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .............................................................TGACTTTGTAGGAAGCCAGAGAGAAG................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................TATGCTGTGCCCTCGGAG................ | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..................................................................TTGTAGGAAGCCAGAGAGAAGACAACAG............................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................CCTCAGGCTGTGCTTGTCT........................................................ | 19 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................TTGCCATCTGTCACAGTCAGGATGTG............................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................TATCAGACATTTCCCGGGGTGGGGTGC.............................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ................................................................CTTTGTAGGAAGCCAGAGAGAAGACAAC.............................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................TGTAGGAAGCCAGAGAGAAGACAACAGTt.......................................................................................................................................................... | 29 | t | 1.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................TGACTTTGTAGGAAGCCAGAGAGAAGACAACAG............................................................................................................................................................ | 33 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| TCTGGATGAGGAGAGCAAAGGACTCGTCCCCTCCTCTCAGTGTCCTCACCCATGTCACTGCTGACTTTGTAGGAAGCCAGAGAGAAGACAACAGTCCTGTTGCCATCTGTCACAGTCAGGATGTGGCCACAATGGCTTCTGGATATATCAGACATTTCCCGGGGTGGGGTGCTGTCCTCAGGCTGTGCTTGTCTCTGCAGAGGTGTCCTGGCCCCCTATGCTGTGCCCTCGGAGCTCCTGCTGGTGGAGG ...........................................................................((.(((((.(((...((((.((((..(((((..((((((.......))))))....)))))....(((((((((...(((((....))))).)))).))))).)))))))).))).))))))).................................................... ................................................................65.....................................................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesWT4() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................CCTCTCAGTGTCCTCAcgc.......................................................................................................................................................................................................... | 19 | cgc | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |