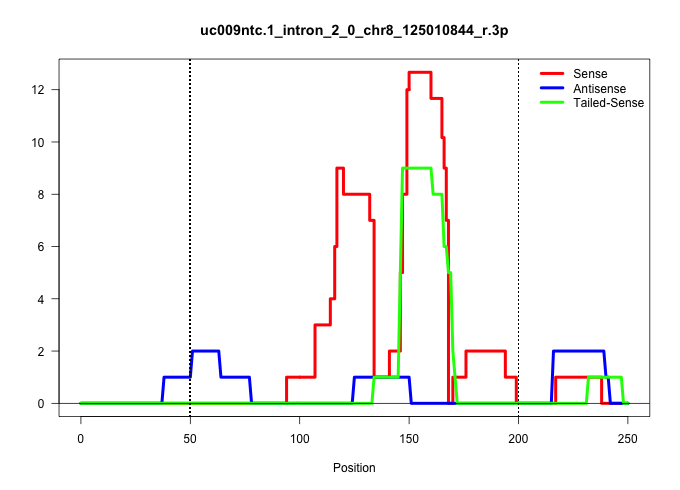
| Gene: BC039210 | ID: uc009ntc.1_intron_2_0_chr8_125010844_r.3p | SPECIES: mm9 |
 |
|
(4) OTHER.mut |
(3) PIWI.ip |
(2) PIWI.mut |
(16) TESTES |
| TCATCATCCTTAACCACATGGTGACAGCCTCGGCTGCCTCCCTGGTGCTGCCCGTGCTTGTGTTCCTGTGGGCCATGCTGACCATCCCGAGGCCTAGCAAGCGCTTTTGGATGACAGCTATCGTCTTCACTGAGGTGGGTAGCAGCTTGTAGGGTAGGGGCTGGGCCACATGGCCCATGGCTGAGTGCCCTGTCCTGCAGGTCATGGTGGTCACCAAATACCTGTTCCAGTTCGGCTTCTTCCCCTGGAA |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT2() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................................................TGTAGGGTAGGGGCTGGGCCAta................................................................................ | 23 | ta | 3.00 | 2.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................TAGGGTAGGGGCTGGGCCA.................................................................................. | 19 | 1 | 3.00 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................CTATCGTCTTCACTGAG.................................................................................................................... | 17 | 1 | 3.00 | 3.00 | - | - | 1.00 | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - |
| ..................................................................................................................................................TTGTAGGGTAGGGGCTGaaa.................................................................................... | 20 | aaa | 2.00 | 0.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................TGTAGGGTAGGGGCTGGGCCA.................................................................................. | 21 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................TTGTAGGGTAGGGGCTGGGCCA.................................................................................. | 22 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................GCTATCGTCTTCACTGAG.................................................................................................................... | 18 | 1 | 2.00 | 2.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................TGGATGACAGCTATCGTCTTCACTG...................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..................................................................................................................CAGCTATCGTCTTCACTGAG.................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................TAGCAAGCGCTTTTGGATGACAGCTA.................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................TTGTAGGGTAGGGGCTGGGCC................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................ATACCTGTTCCAGTTCGGCTT............ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ......................................................................................................................................GTGGGTAGCAGCTTGTAGGGTAGtggc......................................................................................... | 27 | tggc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .....................................................................................................................................................TAGGGTAGGGGCTGGGCC................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................TGGCCCATGGCTGAGTGCCCTGTC........................................................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...........................................................................................................TGGATGACAGCTATCGTCTTCACTGAG.................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................................CGGCTTCTTCCCgctc.. | 16 | gctc | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................GCAGCTTGTAGGGTAGGGGCTGGG..................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................ATGGCTGAGTGCCCTGTCCTGCA................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ......................................................................................................................................GTGGGTAGCAGCTTGTAGGGTAGGGG.......................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..................................................................................................................................................TTGTAGGGTAGGGGCTGGGCCAtttt.............................................................................. | 26 | tttt | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...................................................................................................................................................TGTAGGGTAGGGGCTGGGCCt.................................................................................. | 21 | t | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................TTGTAGGGTAGGGGCTGGGCCAtaa............................................................................... | 25 | taa | 1.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................AGGGTAGGGGCTGGGC.................................................................................... | 16 | 6 | 0.67 | 0.67 | 0.67 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................TGTAGGGTAGGGGCTGGGC.................................................................................... | 19 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 |
| ...................................................................................................................................................TGTAGGGTAGGGGCTGGG..................................................................................... | 18 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - |
| TCATCATCCTTAACCACATGGTGACAGCCTCGGCTGCCTCCCTGGTGCTGCCCGTGCTTGTGTTCCTGTGGGCCATGCTGACCATCCCGAGGCCTAGCAAGCGCTTTTGGATGACAGCTATCGTCTTCACTGAGGTGGGTAGCAGCTTGTAGGGTAGGGGCTGGGCCACATGGCCCATGGCTGAGTGCCCTGTCCTGCAGGTCATGGTGGTCACCAAATACCTGTTCCAGTTCGGCTTCTTCCCCTGGAA |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT2() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................................................................................................TGGCTGAGTGCCCTGTCCTGCAGGTCAa.............................................. | 28 | a | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................TTCACTGAGGTGGGTAGCAGCTTGTA................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................AATACCTGTTCCAGTTCGGCTTCT.......... | 24 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................TCCCTGGTGCTGCCCGTGCTTGTGTT.......................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................AATACCTGTTCCAGTTCGGCTTCTTC........ | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................CCGTGCTTGTGTTCCTGTGGGCCATGC............................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |