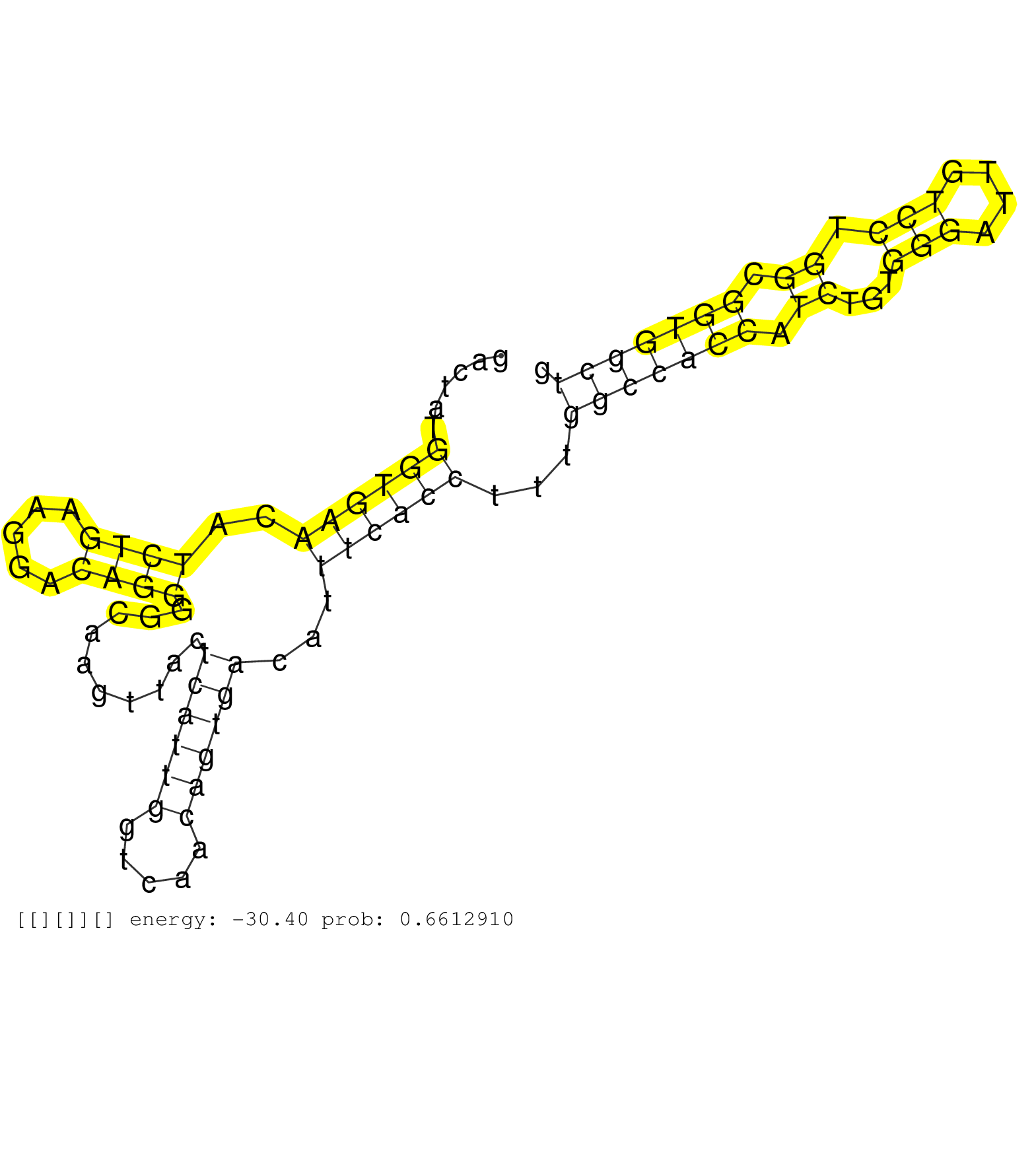

| Gene: Snai3 | ID: uc009nsw.1_intron_0_0_chr8_124978935_r.3p | SPECIES: mm9 |
 |
 |
|
(4) OTHER.mut |
(4) PIWI.ip |
(1) PIWI.mut |
(19) TESTES |
| CCTCTGATGCTGTGATCCTGCTCTGAGTGACAGATGGATTGTCCAAAGGGCTCTGGCCTTGGGTGCTTGGACTATGGTGAACATCTGAAGGACAGGGGCAAGTTACTCATTGGTCAACAGTGACATTTCACCTTTGGCCACCATCTGTGGGATTGTCCTGGCGGTGGCTGCTCGCCTGACCTCCCTTCTTCCTCCCACAGGTGAGAAGCCCTATACCTGTTCTCACTGCAGCAGGGCCTTCGCTGACCGC ...........................................................................((((((..((((.....))))..........((((((.....))))))...))))))...(((((((.((...(((....))).)).)))))))................................................................................. .....................................................................70..................................................................................................170.............................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT1() Testes Data. (testes) | mjTestesWT2() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR051940(GSM545784) 18-32 nt total small RNAs (Mov10l+/-). (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................TGGTGAACATCTGAAGGACAGGGGC....................................................................................................................................................... | 25 | 1 | 5.00 | 5.00 | 4.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................CACCATCTGTGGGATTatg............................................................................................. | 19 | atg | 4.00 | 0.00 | - | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................GGCCTTGGGTGCTTGctgg................................................................................................................................................................................. | 19 | ctgg | 3.00 | 0.00 | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................TGGTGAACATCTGAAGGACAGGGGCA...................................................................................................................................................... | 26 | 1 | 3.00 | 3.00 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................TGAACATCTGAAGGACAGGGGCAAG.................................................................................................................................................... | 25 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .............................................................................TGAACATCTGAAGGACAGGGGCAAGT................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ............................................................................................................................................CCATCTGTGGGATTGTCCTGGCGGTG.................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................TGAAGGACAGGGGCAAGTTACTCATT........................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................TGGACTATGGTGAACATCTGAAGGAC............................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................TGGTGAACATCTGAAGGACAGGGGaat..................................................................................................................................................... | 27 | aat | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...................................................................................TCTGAAGGACAGGGGCAAGTTACTCATT........................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................TGGTGAACATCTGAAGGACAGGGGCAA..................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................TCTCACTGCAGCAGGGCCTTCGCTGA.... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................TGAACATCTGAAGGACAGGGGCAAGTTA................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................GACTATGGTGAACATCTGAAGGACAGG.......................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................ACTATGGTGAACATCTGAAGGACAGGG......................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................TTGTCCAAAGGGCTCTGGCCTTGGGTGC........................................................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..........................................................................TGGTGAACATCTGAAGGACAGGGGCAt..................................................................................................................................................... | 27 | t | 1.00 | 3.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................CCACCATCTGTGGGATca............................................................................................... | 18 | ca | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..........TGTGATCCTGCTCTGAGTGACAGATG...................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................TGGTGAACATCTGAAGGACAGGGG........................................................................................................................................................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................GACTATGGTGAACATCTGAAGGACAG........................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................CCATCTGTGGGATTatgt............................................................................................ | 18 | atgt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................GCCACCATCTGTGGGATT................................................................................................ | 18 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................GCCACCATCTGTGGGATTatg............................................................................................. | 21 | atg | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .......TGCTGTGATCCTGCTCTGAGTGACAGA........................................................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................ACCATCTGTGGGATT................................................................................................ | 15 | 17 | 0.94 | 0.94 | - | 0.18 | - | - | - | - | - | 0.41 | - | - | - | - | - | - | - | 0.18 | 0.12 | - | 0.06 |
| ...........................................................................................................................................ACCATCTGTGGGATTatt............................................................................................. | 18 | att | 0.12 | 0.94 | - | - | - | 0.06 | - | - | - | - | 0.06 | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................CACAGGTGAGAAGCCCTAT.................................... | 19 | 10 | 0.10 | 0.10 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.10 | - |
| CCTCTGATGCTGTGATCCTGCTCTGAGTGACAGATGGATTGTCCAAAGGGCTCTGGCCTTGGGTGCTTGGACTATGGTGAACATCTGAAGGACAGGGGCAAGTTACTCATTGGTCAACAGTGACATTTCACCTTTGGCCACCATCTGTGGGATTGTCCTGGCGGTGGCTGCTCGCCTGACCTCCCTTCTTCCTCCCACAGGTGAGAAGCCCTATACCTGTTCTCACTGCAGCAGGGCCTTCGCTGACCGC ...........................................................................((((((..((((.....))))..........((((((.....))))))...))))))...(((((((.((...(((....))).)).)))))))................................................................................. .....................................................................70..................................................................................................170.............................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT1() Testes Data. (testes) | mjTestesWT2() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | SRR051940(GSM545784) 18-32 nt total small RNAs (Mov10l+/-). (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................GCTCTGAGTGACAGATGGATTGTCCA............................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................GCAAGTTACTCATTGGTCAACAGTGACA............................................................................................................................. | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................TCAACAGTGACATTTCACatta....................................................................................................................... | 22 | atta | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |