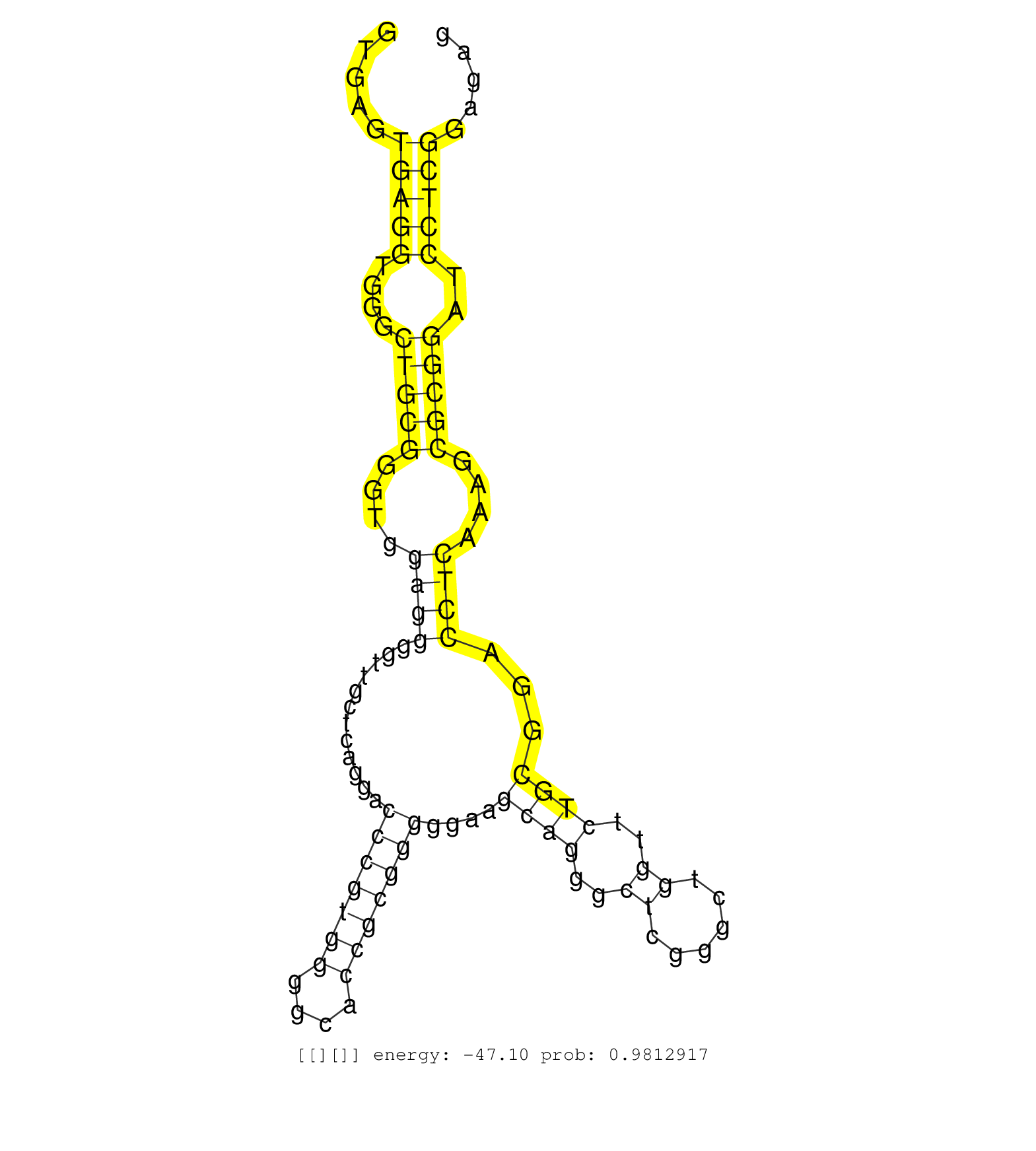

| Gene: Cyba | ID: uc009nsp.1_intron_4_0_chr8_124951625_r.5p | SPECIES: mm9 |
 |
 |
|
(4) OTHER.mut |
(6) PIWI.ip |
(2) PIWI.mut |
(19) TESTES |
| GATCGAGTGGGCCATGTGGGCCAACGAACAGGCGCTGGCGTCTGGCCTGAGTGAGTGAGGTGGGCTGCGGGTGGAGGGGTTGCTCAGGACCCGTGGGGCACCGCGGGGGAAGCAGGGCTCGGGCTGGTTCTGCGGACCTCAAAGCGCGGATCCTCGGAGAGACCTACCGGGGCCTCCGGGCACAGGACGGGCGGTGGCCGCTGCAGGCCGTGCTCGGCCGCCTGTTCTAGAGGCGACCCCGCCCTGCACC .......................................................(((((....(((((....((((............(((((((....)))))))....((((..((......))..))))...))))....)))))..))))).............................................................................................. ..................................................51............................................................................................................161....................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesWT4() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesWT3() Testes Data. (testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM475281(GSM475281) total RNA. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................TGCGGACCTCAAAGCGCGGATCCTCGG............................................................................................. | 27 | 1 | 19.00 | 19.00 | 7.00 | 4.00 | - | 3.00 | 1.00 | 2.00 | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - |
| ..............TGTGGGCCAACGAACAGGCGCTGGCGTC................................................................................................................................................................................................................ | 28 | 1 | 8.00 | 8.00 | - | 3.00 | - | 1.00 | - | 3.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..............TGTGGGCCAACGAACAGGCGCTGGCGT................................................................................................................................................................................................................. | 27 | 1 | 4.00 | 4.00 | - | - | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...........................................................GTGGGCTGCGGGTGGga.............................................................................................................................................................................. | 17 | ga | 4.00 | 0.00 | - | - | - | - | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................TGCGGACCTCAAAGCGCGGATCCTCGGA............................................................................................ | 28 | 1 | 3.00 | 3.00 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................TGCGGACCTCAAAGCGCGGATCCTCG.............................................................................................. | 26 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ......................AACGAACAGGCGCTGGCGTCTGGCCTGAt....................................................................................................................................................................................................... | 29 | t | 2.00 | 0.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........GCCATGTGGGCCAACGAACAGGCGCT...................................................................................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................GTGGGCTGCGGGTGGgg.............................................................................................................................................................................. | 17 | gg | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............TGTGGGCCAACGAACAGGCGCTGGCGTt................................................................................................................................................................................................................ | 28 | t | 1.00 | 4.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............TGTGGGCCAACGAACAGGCGCTGGCGTtt............................................................................................................................................................................................................... | 29 | tt | 1.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .................................GCTGGCGTCTGGCCTGAtt...................................................................................................................................................................................................... | 19 | tt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ............................................................................................................................TGGTTCTGCGGACCTCAAAGCGCGG..................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................TGAGTGAGTGAGGTGGGCTGCGGGT.................................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................TCTGCGGACCTCAAAGCGCGGATCCTCGG............................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............TGTGGGCCAACGAACAGGCGCTGGCGgc................................................................................................................................................................................................................ | 28 | gc | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................TACCGGGGCCTCCGGGCACAGGACGG............................................................ | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................CTGGTTCTGCGGACCTCAAAGCGCGGA.................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................TGCGGACCTCAAAGCGCGGATCCTCGtt............................................................................................ | 28 | tt | 1.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............TGTGGGCCAACGAACAGGCGCTGGCG.................................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............ATGTGGGCCAACGAACAGG.......................................................................................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...........................................................GTGGGCTGCGGGTGGgata............................................................................................................................................................................ | 19 | gata | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..........................................................................................................................................TCAAAGCGCGGATCCTCGGAGAGACC...................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................TGGCGTCTGGCCTGAGTGAGTGAGGca............................................................................................................................................................................................ | 27 | ca | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| GATCGAGTGGGCCATGTGGGCCAACGAACAGGCGCTGGCGTCTGGCCTGAGTGAGTGAGGTGGGCTGCGGGTGGAGGGGTTGCTCAGGACCCGTGGGGCACCGCGGGGGAAGCAGGGCTCGGGCTGGTTCTGCGGACCTCAAAGCGCGGATCCTCGGAGAGACCTACCGGGGCCTCCGGGCACAGGACGGGCGGTGGCCGCTGCAGGCCGTGCTCGGCCGCCTGTTCTAGAGGCGACCCCGCCCTGCACC .......................................................(((((....(((((....((((............(((((((....)))))))....((((..((......))..))))...))))....)))))..))))).............................................................................................. ..................................................51............................................................................................................161....................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesWT4() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesWT3() Testes Data. (testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM475281(GSM475281) total RNA. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................GCTCGGGCTGGTTCTGCGGACCTCAAA........................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................GGGGCACCGCGGGGgtcg.............................................................................................................................................. | 18 | gtcg | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |