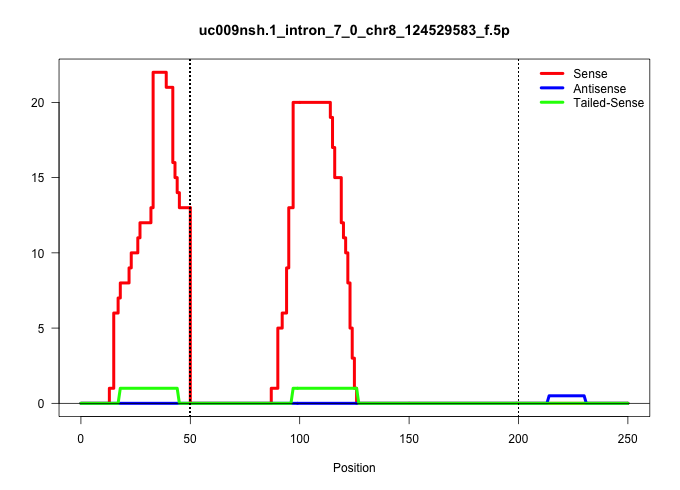
| Gene: Banp | ID: uc009nsh.1_intron_7_0_chr8_124529583_f.5p | SPECIES: mm9 |
 |
|
(1) OVARY |
(4) PIWI.ip |
(3) PIWI.mut |
(16) TESTES |
| CCCAGCAGGTCCAGATCCACCAGATTGGGGAGGATGGACAGGTGCAAGTAGTACGTATCCCGTTACTCCATGGCGGGGGGGGGGGGGTGCTGTGTTCTGAGGCGTAGACAGTGCAGAGGCCAAGCAGTCTTGCACCCTCTGAGCAGACAGCGGGAAGCCGCACCTCAGAAGCTGCTCAAGAGCGGGACTTGGGGGCGGCACTGGGGAGCTGGAGCTTTTAGTCCACCGTGCTGTGGGGCTGTTTTAAAGG |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesWT4() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................ATGGACAGGTGCAAGTA........................................................................................................................................................................................................ | 17 | 1 | 11.00 | 11.00 | - | 6.00 | - | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...............TCCACCAGATTGGGGAGGATGGACAGG................................................................................................................................................................................................................ | 27 | 1 | 4.00 | 4.00 | - | - | 1.00 | 1.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...............................................................................................TCTGAGGCGTAGACAGTGCAGAGGCCA................................................................................................................................ | 27 | 1 | 2.00 | 2.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................TGTGTTCTGAGGCGTAGACAGTGCA....................................................................................................................................... | 25 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .................................................................................................TGAGGCGTAGACAGTGCAGAGGCCAA............................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .................................................................................................TGAGGCGTAGACAGTGCAGAGGCCAAG.............................................................................................................................. | 27 | 1 | 2.00 | 2.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................TGAGGCGTAGACAGTGCAGAGGCCA................................................................................................................................ | 25 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................TGTGTTCTGAGGCGTAGACAGTGCAG...................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - |
| .................................................................................................TGAGGCGTAGACAGTGCAGAGGCCAAGC............................................................................................................................. | 28 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - |
| ..............................................................................................TTCTGAGGCGTAGACAGTGCAGAGG................................................................................................................................... | 25 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................TGTTCTGAGGCGTAGACAGTGCAGAGG................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................GATGGACAGGTGCAAGTA........................................................................................................................................................................................................ | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...........................GGGAGGATGGACAGGTGCAAGTA........................................................................................................................................................................................................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..................ACCAGATTGGGGAGGATGGACAGGTG.............................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ......................GATTGGGGAGGATGGACAGGTGCAAGTA........................................................................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ...............TCCACCAGATTGGGGAGGATGGACAGGT............................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................TCTGAGGCGTAGACAGTGCAGAGGCC................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................ATTGGGGAGGATGGACAGGTGC............................................................................................................................................................................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .................................................................................................TGAGGCGTAGACAGTGCAGAGGCCAAGCA............................................................................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................TCTGAGGCGTAGACAGTGCAGAGGCCAA............................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................ACCAGATTGGGGAGGATGGACAGGTGt............................................................................................................................................................................................................. | 27 | t | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .............GATCCACCAGATTGGGGAGGATGGAC................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................TTCTGAGGCGTAGACAGTGCAGAGGC.................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..........................GGGGAGGATGGACAGGTGCAAGTA........................................................................................................................................................................................................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .......................................................................................TGCTGTGTTCTGAGGCGTAGACAGTGC........................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................TGAGGCGTAGACAGTGCAGAGGCCAAGCAa........................................................................................................................... | 30 | a | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .................CACCAGATTGGGGAGGATGGACAGG................................................................................................................................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| CCCAGCAGGTCCAGATCCACCAGATTGGGGAGGATGGACAGGTGCAAGTAGTACGTATCCCGTTACTCCATGGCGGGGGGGGGGGGGTGCTGTGTTCTGAGGCGTAGACAGTGCAGAGGCCAAGCAGTCTTGCACCCTCTGAGCAGACAGCGGGAAGCCGCACCTCAGAAGCTGCTCAAGAGCGGGACTTGGGGGCGGCACTGGGGAGCTGGAGCTTTTAGTCCACCGTGCTGTGGGGCTGTTTTAAAGG |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesWT4() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................GCTGTGTTCTGAGGCGTAGACAGTGCA....................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ....................................................................................................................................................................................................................................GTGGGGCTGTTTTAgatc.... | 18 | gatc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ......................................................................................................................................................................................................................CTTTTAGTCCACCGTGC................... | 17 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - |