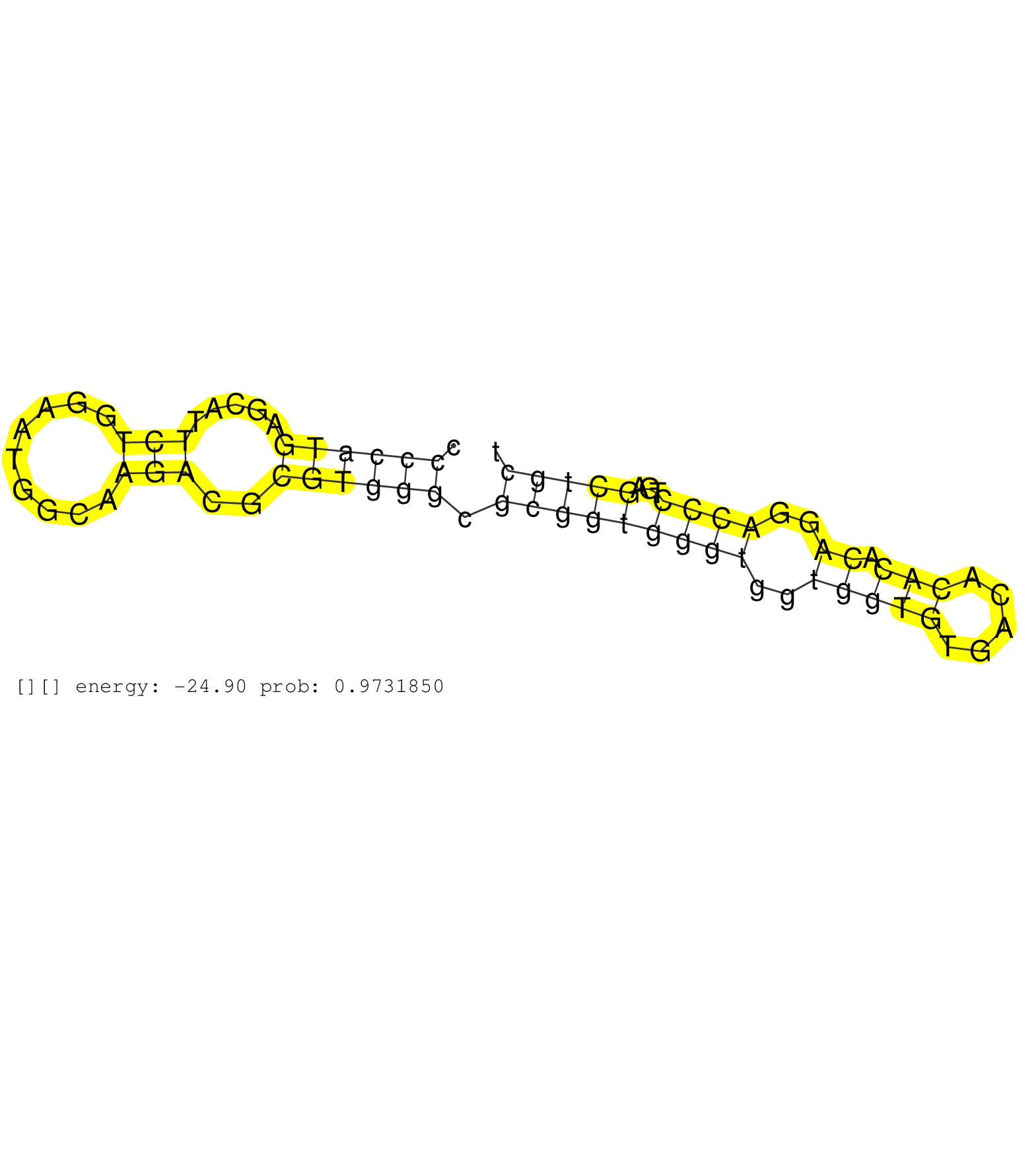

| Gene: Gse1 | ID: uc009nqz.1_intron_1_0_chr8_123077703_f.3p | SPECIES: mm9 |
 |
 |
|
(4) OTHER.mut |
(5) PIWI.ip |
(19) TESTES |
| AGTGTAAAAATAGCTCTCCACCTCCACGCCTGAGAGGCAGTATGACAAGGGAGGCCACAGAATGGCTGTGGGTCCGTCCCTGCCTGCCCGCTCTTGGGCCTCACTCCCCATGAGCATTCTGGAATGGCAAGACGCGTGGGCGCGGTGGGTGGTGGTGTGACACACACAGGACCCTGAGCTGCTTCGTTCTCTTTCCGCAGGGTCCTCACTGAGCAGCGAGTCATCTCCAGTGTCCTCTCCAGCCACCAAC ..........................................................................................................((((((.....(((.........)))..)))))).(((((((((..(((((.....))).))..))))...))))).................................................................... .........................................................................................................106..........................................................................183................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | mjTestesWT1() Testes Data. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesWT2() Testes Data. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................TGAGCATTCTGGAATGGCAAGACGCGT................................................................................................................. | 27 | 1 | 68.00 | 68.00 | 28.00 | 23.00 | 6.00 | 2.00 | 1.00 | - | 3.00 | 1.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 |
| ..............................................................................................................TGAGCATTCTGGAATGGCAAGACGCGTG................................................................................................................ | 28 | 1 | 7.00 | 7.00 | 3.00 | 2.00 | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................GAGCATTCTGGAATGGCAAGACGCGT................................................................................................................. | 26 | 1 | 6.00 | 6.00 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................TGAGCATTCTGGAATGGCAAGACGCG.................................................................................................................. | 26 | 1 | 4.00 | 4.00 | 3.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................ATGAGCATTCTGGAATGGCAAGACGCGT................................................................................................................. | 28 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................TCACTGAGCAGCGAGTCATCT........................ | 21 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...........................................................................................................................................................TGTGACACACACAGGACCCTGAGa....................................................................... | 24 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................ACCCTGAGCTGCTTCGTTCTCTTTCCGCAGa................................................. | 31 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ............................................................................................................................TGGCAAGACGCGTGGGCGCGGTGGG..................................................................................................... | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................CGGTGGGTGGTGGTGcatg......................................................................................... | 19 | catg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .........................................................................................................................................................................................................................GAGTCATCTCCAGTGTCCT.............. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................TGAGCATTCTGGAATGGCAAGACGagt................................................................................................................. | 27 | agt | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................TGAGAGGCAGTATGACAAGGGAG..................................................................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................TGACACACACAGGACCCTGAG........................................................................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................TGAGAGGCAGTATGACAAGGGAGGC................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................TGGAATGGCAAGACGCGTGGGCGCGGgg....................................................................................................... | 28 | gg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .......................................................................................................................TGGAATGGCAAGACGCGTGGGCGCGGT........................................................................................................ | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................TGAGCAGCGAGTCATCTCCAGT................... | 22 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................TGGTGTGACACACACAGGACCCTGAGC....................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .........................................................................................................CCCCATGAGCATTCTGGAATGGCAAGA...................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................TGAGCATTCTGGAATGGCAAGACGt................................................................................................................... | 25 | t | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................AGCATTCTGGAATGGCAAGACGCGT................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................TGGGCGCGGTGGGTGGTGG............................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..........................................TGACAAGGGAGGCCACAGAAT........................................................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................TGAGCAGCGAGTCATCTCCAGTGTCCTC............. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| AGTGTAAAAATAGCTCTCCACCTCCACGCCTGAGAGGCAGTATGACAAGGGAGGCCACAGAATGGCTGTGGGTCCGTCCCTGCCTGCCCGCTCTTGGGCCTCACTCCCCATGAGCATTCTGGAATGGCAAGACGCGTGGGCGCGGTGGGTGGTGGTGTGACACACACAGGACCCTGAGCTGCTTCGTTCTCTTTCCGCAGGGTCCTCACTGAGCAGCGAGTCATCTCCAGTGTCCTCTCCAGCCACCAAC ..........................................................................................................((((((.....(((.........)))..)))))).(((((((((..(((((.....))).))..))))...))))).................................................................... .........................................................................................................106..........................................................................183................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | mjTestesWT1() Testes Data. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesWT2() Testes Data. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................CCTCACTCCCCATGAGCATTCTGGAA.............................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................CAAGGGAGGCCACAGagcc.............................................................................................................................................................................................. | 19 | agcc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |