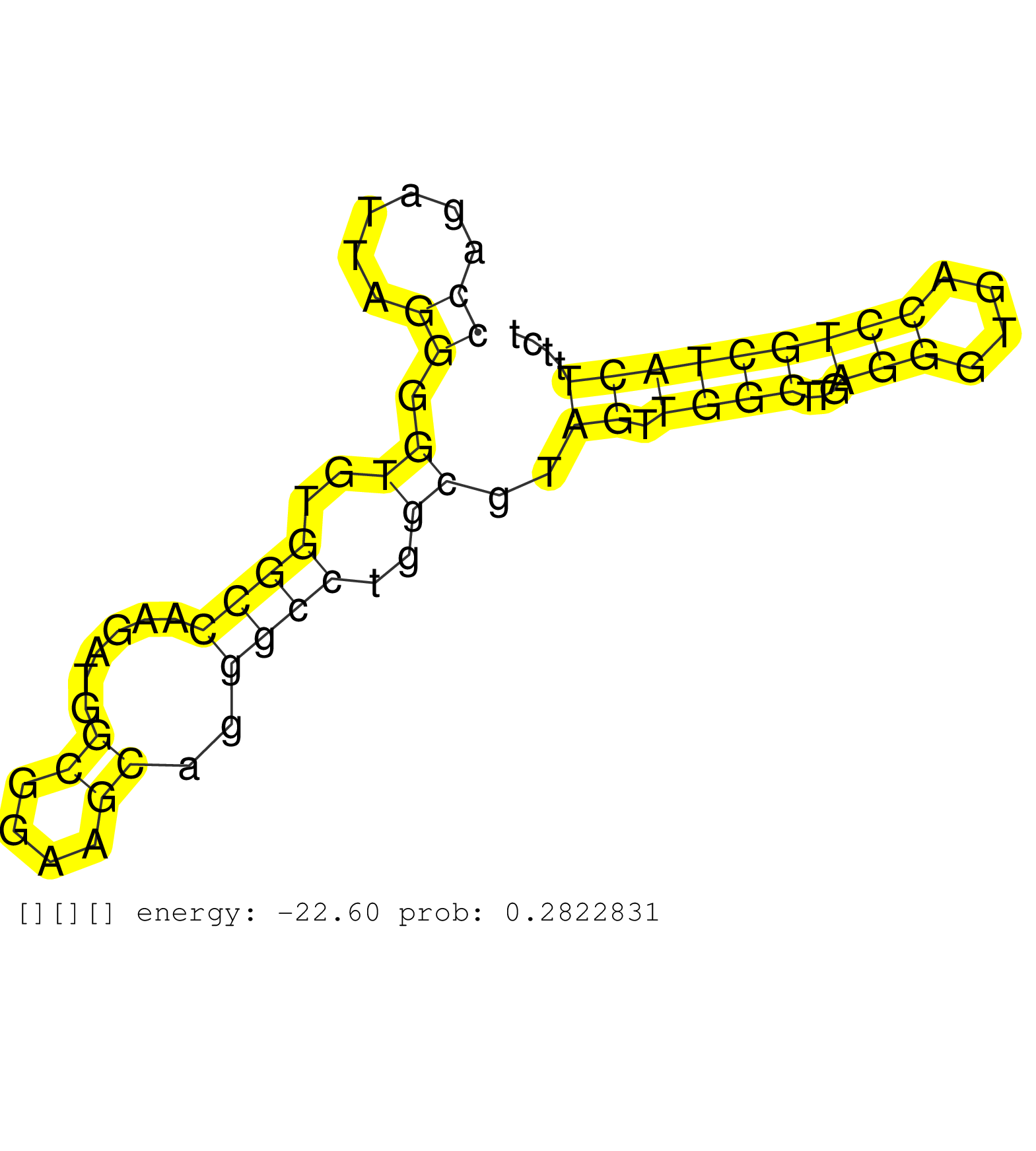

| Gene: 1700030J22Rik | ID: uc009nor.1_intron_0_0_chr8_119496057_r.3p | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(5) PIWI.ip |
(1) PIWI.mut |
(15) TESTES |
| CAGGAGTTATGGCCAGTGAGGCTACATAAGACCGTGTGTGTGTGTGTGTGTGTGTATGTGTGTGTGTGTATTTAAGATAAGTCAAGTTTAAGAAAAAAAACATGGCTCAAGTAAGGCTACCAGATTAGGGGTGTGGCCAAGATGGCGGAAGCAGGGCCTGGCGTAGTTGGCTTGAGGGTGACCTGCTACTTTCTCTGCAGGTCCATGGCTGGGGAAGGACCTCACCAACTGCCTGCATCTGGTCAAAGAA .......................................................................................................................((......)).((..((((......((....))..))))..))..((.((((...(((....)))))))))............................................................ .......................................................................................................................120.......................................................................194...................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................................................................................TACTTTCTCTGCAGGTCCATGGCTGG...................................... | 26 | 1 | 6.00 | 6.00 | 1.00 | 2.00 | - | - | - | - | - | 2.00 | 1.00 | - | - | - | - | - | - |
| ............................................................................................................................TTAGGGGTGTGGCCAAGATGGCGGAAGC.................................................................................................. | 28 | 1 | 3.00 | 3.00 | - | - | - | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................TACCAGATTAGGGGTGTGGCCAAGA............................................................................................................ | 25 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - |
| .....................................................................................................................TACCAGATTAGGGGTGTGGCCAAGAT........................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .......................................................................................................................................................................................TGCTACTTTCTCTGCAGGTCCATGGC......................................... | 26 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................GATTAGGGGTGTGGCCAAGATGGCGG...................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ............................................................................................................................TTAGGGGTGTGGCCAAGATGGCGGAAGCA................................................................................................. | 29 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................TAGTTGGCTTGAGGGTGACCTGCTACT............................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..............................................................TGTGTGTATTTAAGATAAGTCAAGTTT................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ........................................................................................TAAGAAAAAAAACATGGCTCAAGTAA........................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ............................................................................................................AAGTAAGGCTACCAttcc............................................................................................................................ | 18 | ttcc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .........................................................GTGTGTGTGTGTATTTAAGATAAGTCAAGTT.................................................................................................................................................................. | 31 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................TAGGGGTGTGGCCAAGATGGCGGAAG................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................TGTGTGTGTGTATTTAAGATAAGTCA...................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................TACTTTCTCTGCAGGTCCATGGCTG....................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................TGGCGTAGTTGGCTTGAGGGTGACa................................................................... | 25 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ......................................................................................................................ACCAGATTAGGGGTGTGGCCAAGAT........................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................GTAGTTGGCTTGAGGGTGACCTGCaa.............................................................. | 26 | aa | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .........TGGCCAGTGAGGCTACATAAGACCGTGT..................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................TGTGTGTGTGTATTTAAGATAAGTCAAGT................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..............................................................................................AAAAAACATGGCTCAAGTAAGGCTACC................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ......................................................................TTTAAGATAAGTCAAGTTTAAGAAAAAAAt...................................................................................................................................................... | 30 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .................................................................................................................................................................................................TCTGCAGGTCCATGGCTGGGGAAGG................................ | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................TGTGTGTGTGTGTATTTAAGATAAGTC....................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................CAGATTAGGGGTGTGGCCAAGATGG......................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................TGTGTGTATTTAAGATAAGTCAAGTT.................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................TGTGTGTGTATTTAAGATAAGTCAAGTT.................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ......................................................................................................TGGCTCAAGTAAGGCTACCAGATTAG.......................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................TTAGGGGTGTGGCCAAGATGGCGGAAG................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................ATAAGACCGTGTGTGTGTGTGTGTG........................................................................................................................................................................................................ | 25 | 4 | 0.25 | 0.25 | - | - | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - |
| CAGGAGTTATGGCCAGTGAGGCTACATAAGACCGTGTGTGTGTGTGTGTGTGTGTATGTGTGTGTGTGTATTTAAGATAAGTCAAGTTTAAGAAAAAAAACATGGCTCAAGTAAGGCTACCAGATTAGGGGTGTGGCCAAGATGGCGGAAGCAGGGCCTGGCGTAGTTGGCTTGAGGGTGACCTGCTACTTTCTCTGCAGGTCCATGGCTGGGGAAGGACCTCACCAACTGCCTGCATCTGGTCAAAGAA .......................................................................................................................((......)).((..((((......((....))..))))..))..((.((((...(((....)))))))))............................................................ .......................................................................................................................120.......................................................................194...................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) |
|---|