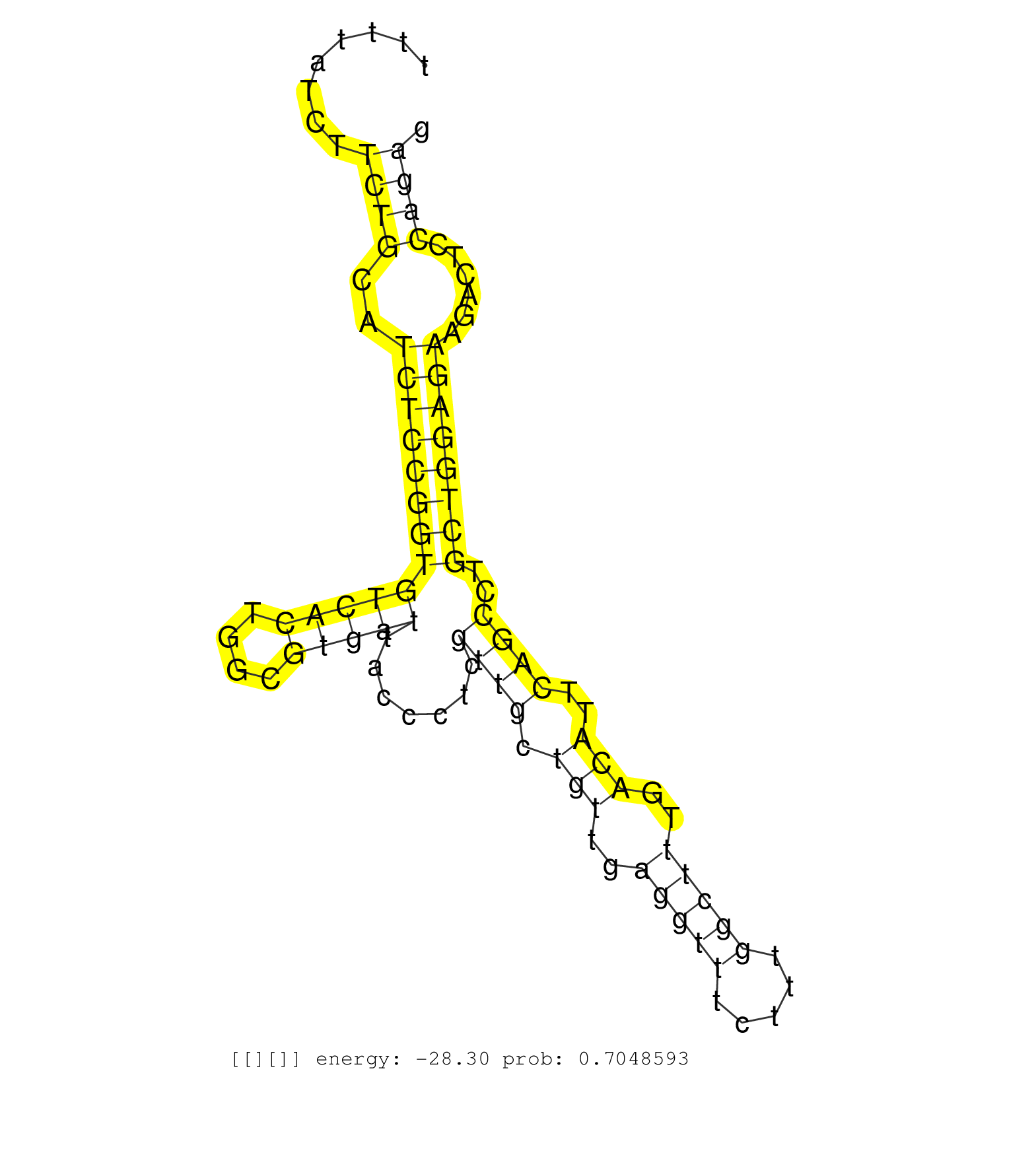

| Gene: AK020517 | ID: uc009nlx.1_intron_2_0_chr8_113667625_r.3p | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(5) PIWI.ip |
(18) TESTES |
| TTACTCTCAAGGACTTTTTAAAGGGTGTCCATTGCTTTAAGCTTACTGCCTTTTATCTTCTGCATCTCCGGTGTCACTGGCGTGATTACCCTCGTTGCTGTTGAGGTTTCTTTGGCTTTGACATTCAGCCTGCTGGAGAAGACTCCAGAGCCTCCCTTTGCTTCTTGCTATTTGGTTTAATCCCTTCTGCTCTCACCTAGTCACTGTTGTAGGTCAAGATGAACACTAAACCAACAGTCTGTCAGGCTGG ..........................................................((((..(((((((((((((....))))).......((((.(((..(((((.....)))))..)))..))))..))))))))......))))..................................................................................................... ..................................................51.................................................................................................150.................................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | mjTestesWT2() Testes Data. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................TTAAAGGGTGTCCATTGCTTTAAGCTT.............................................................................................................................................................................................................. | 27 | 1 | 6.00 | 6.00 | 2.00 | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................TGACATTCAGCCTGCTGGAGAAGACTCC........................................................................................................ | 28 | 1 | 5.00 | 5.00 | - | - | - | - | - | - | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 |
| .TACTCTCAAGGACTTTTTAAAGGGTG............................................................................................................................................................................................................................... | 26 | 1 | 5.00 | 5.00 | - | 2.00 | - | - | - | - | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................TAGTCACTGTTGTAGGTCAAGATGAAC.......................... | 27 | 1 | 4.00 | 4.00 | - | 1.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...............................................................................................................................................................................................................TGTAGGTCAAGATGAACACTAAACCAAC............... | 28 | 1 | 3.00 | 3.00 | - | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................TGACATTCAGCCTGCTGGAGAAGACTC......................................................................................................... | 27 | 1 | 3.00 | 3.00 | - | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................TGACATTCAGCCTGCTGGAGAAGACT.......................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................TGAACACTAAACCAACAGTCTGTCAGGCT.. | 29 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .TACTCTCAAGGACTTTTTAAAGGGTGTC............................................................................................................................................................................................................................. | 28 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................TCTTCTGCATCTCCGGTGTCACTGGCG........................................................................................................................................................................ | 27 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................TGTTGTAGGTCAAGATGAACACTAAAC................... | 27 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................TAGGTCAAGATGAACACTAAACCAACAGTC........... | 30 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................TTACTGCCTTTTATCTTCTGCATCTCC..................................................................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................TTTTATCTTCTGCATCTCCGGTGTC............................................................................................................................................................................... | 25 | 1 | 2.00 | 2.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................TGACATTCAGCCTGCTGGAGAAGAatcc........................................................................................................ | 28 | atcc | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................TGCCTTTTATCTTCTGCATCTCCGGT.................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................TGCATCTCCGGTGTCACTGGCGT....................................................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................TGACATTCAGCCTGCTGGAGAAGAC........................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................TTGACATTCAGCCTGCTGGAGAAGACT.......................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............TTTTTAAAGGGTGTCCATTG........................................................................................................................................................................................................................ | 20 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................TGGCGTGATTACCCTCGTTGCTGTTG................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................TAGGTCAAGATGAACACTAAACCAACAG............. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................TGCTGTTGAGGTTTCTTTGGCTTTtac................................................................................................................................ | 27 | tac | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................GATGAACACTAAACCAACAGTCTGTCAGG.... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................TTTTATCTTCTGCATCTCCGGTGTCA.............................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ................TTTAAAGGGTGTCCATTG........................................................................................................................................................................................................................ | 18 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................GATGAACACTAAACCAACAGTCTGTCAGGC... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................CCTAGTCACTGTTGcatt..................................... | 18 | catt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ......................................................................................................................TGACATTCAGCCTGCTGGAGAAGACTCCA....................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................TTTAAAGGGTGTCCATTGCTTTAAGCTT.............................................................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................TAAAGGGTGTCCATTGCTTTAAGCTTAC............................................................................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................TATCTTCTGCATCTCCGGTGTCACTGGC......................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ....................................................................................................................TTTGACATTCAGCCTGCTGGAGAAGAC........................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................TAGTCACTGTTGTAGGTCAAGATGAAa.......................... | 27 | a | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............TTTTAAAGGGTGTCCATTGCTTTAAcctt.............................................................................................................................................................................................................. | 29 | cctt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .....................................................................................................................................................................................................TAGTCACTGTTGTAGGTCAAGATGAA........................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................TTCTGCATCTCCGGTGTCACTGGC......................................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................TCTGCATCTCCGGTGTCA.............................................................................................................................................................................. | 18 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................TGACATTCAGCCTGCTGtat................................................................................................................ | 20 | tat | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ....................................................................................................................................................AGCCTCCCTTTGCTTCTTGCTATTTG............................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................TACTGCCTTTTATCTTCTGCATCTCCGG................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................TGAACACTAAACCAACAGTCTGTCAGGC... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................CCCTCGTTGCTGTTGAGGTTTCTTTGG....................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................TCTTCTGCATCTCCGGTGTCACTGGCGT....................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................TCTGCATCTCCGGTGTCACTGGCGTG...................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ......................................................................................................................TGACATTCAGCCTGCTGtt................................................................................................................. | 19 | tt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..................................................................................................TGTTGAGGTTTCTTTctgt..................................................................................................................................... | 19 | ctgt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...............TTTTAAAGGGTGTCCATTGCTTTAAGCT............................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ................................................................TCTCCGGTGTCACTGGCGTGATTACCC............................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TTACTCTCAAGGACTTTTTAAAGGGTGTCCATTGCTTTAAGCTTACTGCCTTTTATCTTCTGCATCTCCGGTGTCACTGGCGTGATTACCCTCGTTGCTGTTGAGGTTTCTTTGGCTTTGACATTCAGCCTGCTGGAGAAGACTCCAGAGCCTCCCTTTGCTTCTTGCTATTTGGTTTAATCCCTTCTGCTCTCACCTAGTCACTGTTGTAGGTCAAGATGAACACTAAACCAACAGTCTGTCAGGCTGG ..........................................................((((..(((((((((((((....))))).......((((.(((..(((((.....)))))..)))..))))..))))))))......))))..................................................................................................... ..................................................51.................................................................................................150.................................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR014236(GSM319960) 10 dpp total. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM179088(GSM179088) Developmentally regulated piRNA clusters implicate MILI in transposon control. (piwi testes) | mjTestesWT2() Testes Data. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................GCTTTGACATTCAGCgaag......................................................................................................................... | 19 | gaag | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |