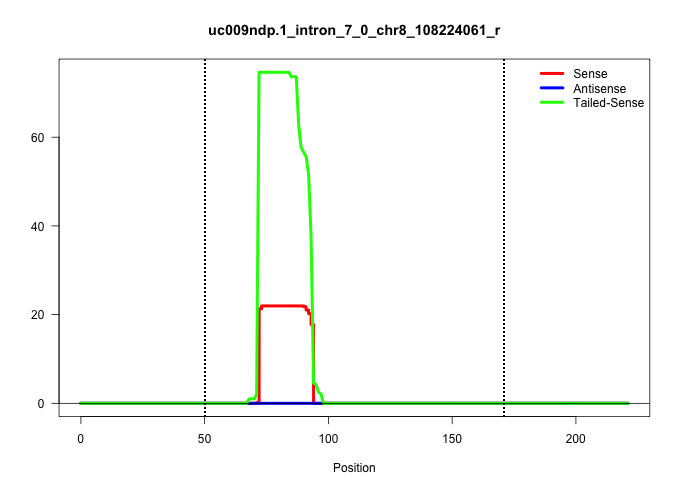
| Gene: Acd | ID: uc009ndp.1_intron_7_0_chr8_108224061_r | SPECIES: mm9 |
 |
 |
|
(1) OTHER.ip |
(6) OTHER.mut |
(4) OVARY |
(2) PIWI.ip |
(2) PIWI.mut |
(1) TDRD1.ip |
(25) TESTES |
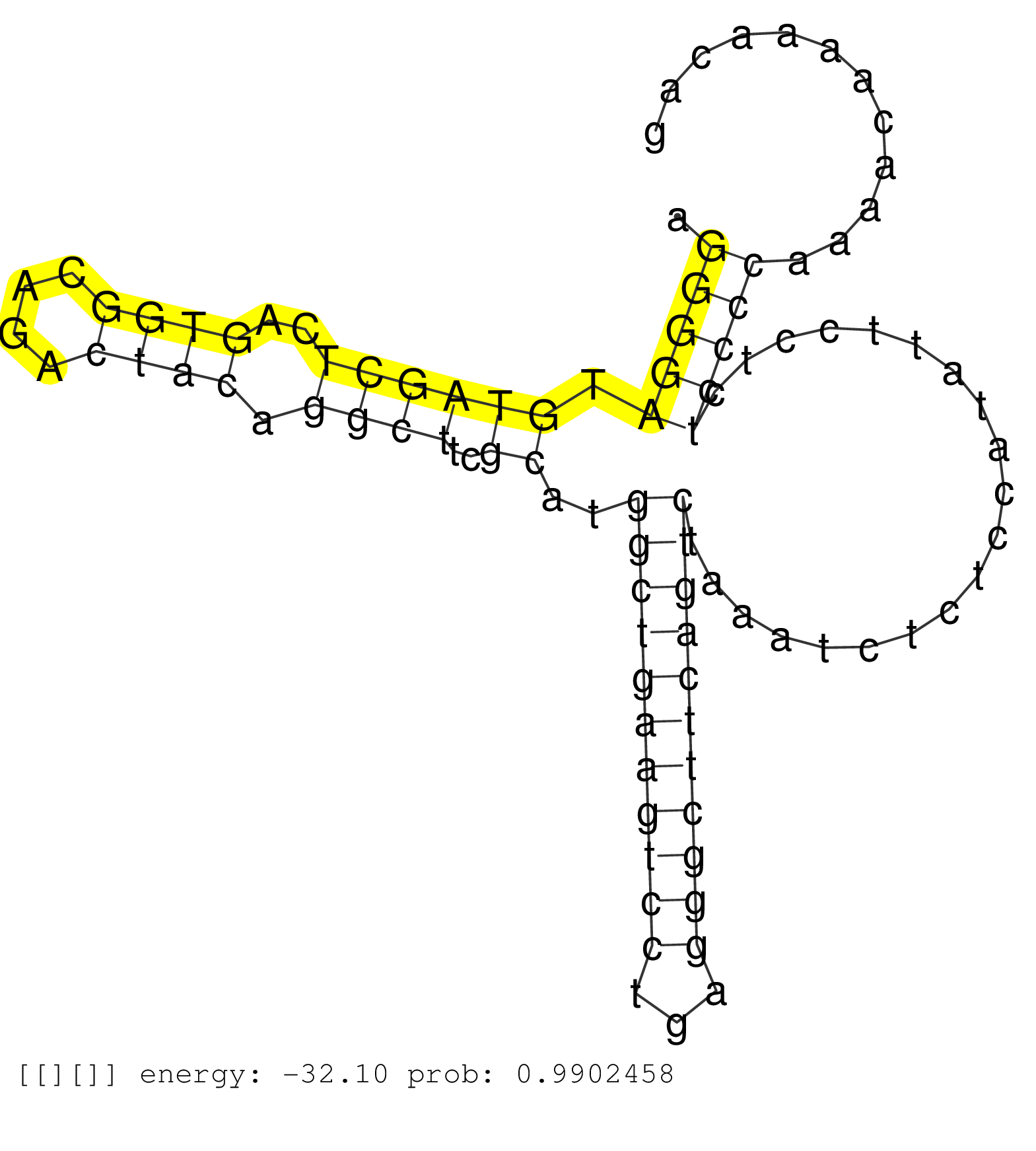
| CGCTTTAACCTGCTGCCCACGGAGCAGCCCCGGATACAGGTGACTGGTTGGTAAGTGGTCCCTTTAAGGGAAGGGGATGTAGCTCAGTGGCAGACTACAGGCTTCGCATGGCTGAAGTCCTGAGGGCTTCAGTCTAAATCTCTCCATATTCCTCTCCCCAAAACAAAACAGTTAAATGAAGCTTCCACCTTTTGTCTCTTCACGCGCACTTGGCATAGATC ........................................................................(((((.((((((..((((....)))).))))..))..(((((((((((...)))))))))))....................))))).............................................................. .......................................................................72.................................................................................................171................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | mjTestesWT1() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR037898(GSM510434) ovary_rep3. (ovary) | SRR037897(GSM510433) ovary_rep2. (ovary) | SRR037902(GSM510438) testes_rep3. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR037899(GSM510435) ovary_rep4. (ovary) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | mjTestesWT3() Testes Data. (testes) | SRR037901(GSM510437) testes_rep2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................GGGGATGTAGCTCAGTGGagaa............................................................................................................................... | 22 | agaa | 20.00 | 0.00 | 13.00 | 5.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................GGGGATGTAGCTCAGTGGCAGA............................................................................................................................... | 22 | 8 | 17.88 | 17.88 | 5.50 | 7.12 | 1.38 | 0.75 | - | 1.12 | - | - | - | - | 0.38 | 0.25 | 0.12 | 0.25 | 0.12 | - | 0.12 | 0.12 | - | - | - | 0.12 | 0.12 | 0.12 | 0.12 | - | 0.12 | - | - |
| ........................................................................GGGGATGTAGCTCtta..................................................................................................................................... | 16 | tta | 10.00 | 0.00 | 2.00 | 2.00 | 4.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................ACAGGTGACTGGTTGGTAAGTGG................................................................................................................................................................... | 23 | 1 | 7.00 | 7.00 | - | - | - | - | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................GGGGATGTAGCTCAGTGGagt................................................................................................................................ | 21 | agt | 5.00 | 0.00 | 3.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................GGGGATGTAGCTCAGTGtaga................................................................................................................................ | 21 | taga | 3.00 | 0.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................GGGGATGTAGCTCAGTGGaagt............................................................................................................................... | 22 | aagt | 3.00 | 0.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................GGGGATGTAGCTCAGTGGCAG................................................................................................................................ | 21 | 11 | 2.64 | 2.64 | 1.09 | 0.73 | 0.18 | 0.18 | - | - | - | - | - | - | - | 0.09 | - | - | 0.09 | 0.09 | - | - | 0.09 | 0.09 | - | - | - | - | - | - | - | - | - |
| ........................................................................GGGGATGTAGCTCAGTGtag................................................................................................................................. | 20 | tag | 2.00 | 0.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................GGGGATGTAGCTCAGTGGaat................................................................................................................................ | 21 | aat | 2.00 | 0.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................GGGGATGTAGCTCAGTGGgagt............................................................................................................................... | 22 | gagt | 2.00 | 0.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................GGGGATGTAGCTCttat.................................................................................................................................... | 17 | ttat | 2.00 | 0.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................GGGGATGTAGCTCAGTGGagat............................................................................................................................... | 22 | agat | 2.00 | 0.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................CCTTTAAGGGAAGGGGATGTAGCTCAG...................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................GGGGATGTAGCTCAGTGGagc................................................................................................................................ | 21 | agc | 2.00 | 0.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................GGGGATGTAGCTCAGTGGagtg............................................................................................................................... | 22 | agtg | 2.00 | 0.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................GGGGATGTAGCTCAGTGttt................................................................................................................................. | 20 | ttt | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................GGGGATGTAGCTCAGTGttat................................................................................................................................ | 21 | ttat | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................GGGGATGTAGCTCAGTGGggaa............................................................................................................................... | 22 | ggaa | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................GGGGATGTAGCTCttaa.................................................................................................................................... | 17 | ttaa | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................GGAAGGGGATGTAagct........................................................................................................................................ | 17 | agct | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................GGGGATGTAGCTCAGTGGagcg............................................................................................................................... | 22 | agcg | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................GGGGATGTAGCTCAGTcgta................................................................................................................................. | 20 | cgta | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................GGGGATGTAGCTCAGTGGgagc............................................................................................................................... | 22 | gagc | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................GGGGATGTAGCTCAGTGGCAGACcgc........................................................................................................................... | 26 | cgc | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................GGGGATGTAGCTCAGTGtgag................................................................................................................................ | 21 | tgag | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................GGGGATGTAGCTCAcagg................................................................................................................................... | 18 | cagg | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................GGGGATGTAGCTCAGTcgt.................................................................................................................................. | 19 | cgt | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................AGGGGATGTAGCTCAaa..................................................................................................................................... | 17 | aa | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................GGGGATGTAGCTCAGTGGatt................................................................................................................................ | 21 | att | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................GGGGATGTAGCTCAGTGGgcga............................................................................................................................... | 22 | gcga | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................GGGGATGTAGCTCtatt.................................................................................................................................... | 17 | tatt | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................GGGGATGTAGCTCtttc.................................................................................................................................... | 17 | tttc | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................GGGGATGTAGCTCAGTGGC.................................................................................................................................. | 19 | 19 | 0.95 | 0.95 | 0.11 | 0.37 | 0.11 | 0.11 | - | - | 0.05 | - | - | - | - | - | - | - | - | - | - | - | - | 0.05 | 0.05 | - | - | - | - | - | - | 0.05 | 0.05 |
| ........................................................................GGGGATGTAGCTCAGTGGCA................................................................................................................................. | 20 | 16 | 0.81 | 0.81 | 0.25 | 0.12 | 0.12 | 0.12 | - | - | - | 0.06 | - | - | - | - | - | - | - | - | 0.06 | - | 0.06 | - | - | - | - | - | - | - | - | - | - |
| .........................................................................GGGATGTAGCTCAGTGGCAGA............................................................................................................................... | 21 | 17 | 0.65 | 0.65 | 0.29 | 0.29 | 0.06 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................GGGGATGTAGCTCAGTGGCAGAgg............................................................................................................................. | 24 | gg | 0.50 | 17.88 | 0.12 | 0.12 | - | - | - | - | - | - | - | - | - | - | 0.12 | - | - | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................GGGGATGTAGCTCAGTGGCAGcgc............................................................................................................................. | 24 | cgc | 0.36 | 2.64 | 0.27 | - | 0.09 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................AGGGGATGTAGCTCAGTGG................................................................................................................................... | 19 | 11 | 0.27 | 0.27 | - | - | - | 0.09 | - | 0.09 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.09 | - | - | - | - | - | - | - | - |
| ........................................................................GGGGATGTAGCTCAGTGGCAGAgaaa........................................................................................................................... | 26 | gaaa | 0.25 | 17.88 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................GGGGATGTAGCTCAGTGGCAGAaa............................................................................................................................. | 24 | aa | 0.25 | 17.88 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................GGGGATGTAGCTCAGTGGCAGAtt............................................................................................................................. | 24 | tt | 0.25 | 17.88 | - | - | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................GGGGATGTAGCTCAGTGGCAGAgcgc........................................................................................................................... | 26 | gcgc | 0.25 | 17.88 | - | - | - | - | - | - | - | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.12 | - | - | - |
| ........................................................................GGGGATGTAGCTCAGTGGCAGAt.............................................................................................................................. | 23 | t | 0.12 | 17.88 | - | - | - | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................GGGGATGTAGCTCAGTGGCAGAgcag........................................................................................................................... | 26 | gcag | 0.12 | 17.88 | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................GGGGATGTAGCTCAGTGGCAGAat............................................................................................................................. | 24 | at | 0.12 | 17.88 | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................GGGGATGTAGCTCAGTGGCAGAgct............................................................................................................................ | 25 | gct | 0.12 | 17.88 | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................GGGGATGTAGCTCAGTGGCAGAta............................................................................................................................. | 24 | ta | 0.12 | 17.88 | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................GGGGATGTAGCTCAGTGGCAGAgta............................................................................................................................ | 25 | gta | 0.12 | 17.88 | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................GGGGATGTAGCTCAGTGGCAGAgga............................................................................................................................ | 25 | gga | 0.12 | 17.88 | - | - | - | - | - | - | - | - | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................GGGGATGTAGCTCAGTGGCAGAgcc............................................................................................................................ | 25 | gcc | 0.12 | 17.88 | - | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................GGGGATGTAGCTCAGTGGCAGAgccg........................................................................................................................... | 26 | gccg | 0.12 | 17.88 | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................GGGGATGTAGCTCAGTGGCAGAgagc........................................................................................................................... | 26 | gagc | 0.12 | 17.88 | - | - | - | - | - | - | - | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................GGGGATGTAGCTCAGTGGCAaagc............................................................................................................................. | 24 | aagc | 0.12 | 0.81 | 0.06 | 0.06 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................GGGGATGTAGCTCAGTGGCAGAgctc........................................................................................................................... | 26 | gctc | 0.12 | 17.88 | - | - | - | - | - | - | - | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................GGGGATGTAGCTCAGTGGCAGAgcca........................................................................................................................... | 26 | gcca | 0.12 | 17.88 | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................GGGGATGTAGCTCAGTGGCcgag.............................................................................................................................. | 23 | cgag | 0.11 | 0.95 | - | 0.05 | 0.05 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................GGGGATGTAGCTCAGTGGCAGc............................................................................................................................... | 22 | c | 0.09 | 2.64 | - | - | - | - | - | - | - | - | - | - | 0.09 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................GGGGATGTAGCTCAGTGGCAcag.............................................................................................................................. | 23 | cag | 0.06 | 0.81 | - | - | 0.06 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................GGGGATGTAGCTCAGTGGCgag............................................................................................................................... | 22 | gag | 0.05 | 0.95 | 0.05 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................GGGGATGTAGCTCAGTGGCgagc.............................................................................................................................. | 23 | gagc | 0.05 | 0.95 | - | - | - | - | - | - | - | - | - | - | - | - | 0.05 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................GGGGATGTAGCTCAGTGGCgggc.............................................................................................................................. | 23 | gggc | 0.05 | 0.95 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.05 | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................GGGGATGTAGCTCAGTGGCtacc.............................................................................................................................. | 23 | tacc | 0.05 | 0.95 | - | - | - | - | - | - | - | - | 0.05 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CGCTTTAACCTGCTGCCCACGGAGCAGCCCCGGATACAGGTGACTGGTTGGTAAGTGGTCCCTTTAAGGGAAGGGGATGTAGCTCAGTGGCAGACTACAGGCTTCGCATGGCTGAAGTCCTGAGGGCTTCAGTCTAAATCTCTCCATATTCCTCTCCCCAAAACAAAACAGTTAAATGAAGCTTCCACCTTTTGTCTCTTCACGCGCACTTGGCATAGATC ........................................................................(((((.((((((..((((....)))).))))..))..(((((((((((...)))))))))))....................))))).............................................................. .......................................................................72.................................................................................................171................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | mjTestesWT1() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR037898(GSM510434) ovary_rep3. (ovary) | SRR037897(GSM510433) ovary_rep2. (ovary) | SRR037902(GSM510438) testes_rep3. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR037899(GSM510435) ovary_rep4. (ovary) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR051939(GSM545783) Mov10L1-associated piRNAs. (mov10L testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | mjTestesWT3() Testes Data. (testes) | SRR037901(GSM510437) testes_rep2. (testes) |
|---|