
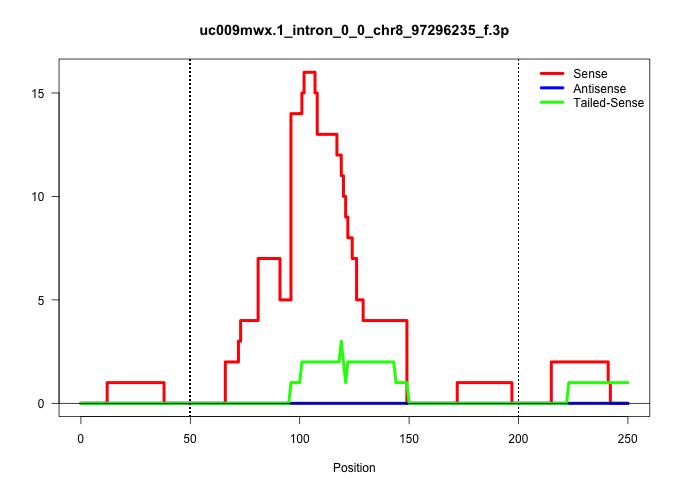
| Gene: Cx3cl1 | ID: uc009mwx.1_intron_0_0_chr8_97296235_f.3p | SPECIES: mm9 |
 |
 |
|
(2) PIWI.ip |
(9) TESTES |
| AAATGAGATTTATAAATAAAGGCTAATTTTTTATCAGTCCAAAACAACCCCAGAGGCTGAGACTCTTGCTGTCTGGCAGGTTATCACGGGTTGGGTTAGATGTGGATTCTGGCTTGGGAGTATAGCCTCGAGGAGCTGAAATGGGTATGGGGCACCAGATGCCCAGGGGTCCTACTGACTGCTGGCCTCTGTGGATCCAGGTCAGCACCTCGGCATGACGAAATGCGAAATCATGTGCGACAAGATGACC ......................................................................................................(((((((.(((.(((.(((.(((.....((((.((....(((((((((....)))..)))))).)).)))).)))))).)))))).....)))))))................................................... ...........................................................................................92..........................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................TAGATGTGGATTCTGGCTTGGGAGTA................................................................................................................................ | 26 | 1 | 5.00 | 5.00 | 3.00 | - | - | 2.00 | - | - | - | - | - |
| ..........................................................................................................................TAGCCTCGAGGAGCTGAAATGGGTATG..................................................................................................... | 27 | 1 | 4.00 | 4.00 | - | - | 1.00 | 1.00 | - | 1.00 | 1.00 | - | - |
| .................................................................................TATCACGGGTTGGGTTAGATGTGGATT.............................................................................................................................................. | 27 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | 1.00 | - | - | - | - |
| ..................................................................TGCTGTCTGGCAGGTTATCACGGGT............................................................................................................................................................... | 25 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | 1.00 | - |
| .....................................................................................................GTGGATTCTGGCTTGGcat.................................................................................................................................. | 19 | cat | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - |
| .....................................................................................................GTGGATTCTGGCTTGGGAGTATAGC............................................................................................................................ | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - |
| ............TAAATAAAGGCTAATTTTTTATCAGT.................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - |
| ................................................................................................TAGATGTGGATTCTGGCTTGGGA................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................TGACGAAATGCGAAATCATGTGCGAC......... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - |
| ..........................................................................................................................TAGCCTCGAGGAGCTGAAATGGGTATGt.................................................................................................... | 28 | t | 1.00 | 4.00 | 1.00 | - | - | - | - | - | - | - | - |
| ................................................................................................TAGATGTGGATTCTGGCTTGGGAGa................................................................................................................................. | 25 | a | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - |
| ................................................................................................TAGATGTGGATTCTGGCTTGG..................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - |
| ........................................................................CTGGCAGGTTATCACGGGTTGGGTTAG....................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - |
| ................................................................................................TAGATGTGGATTCTGGCTTGGGAGT................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - |
| ....................................................................................................TGTGGATTCTGGCTTGGGAGTATAGC............................................................................................................................ | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - |
| .................................................................................TATCACGGGTTGGGTTAGATGTGGAT............................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - |
| .........................................................................TGGCAGGTTATCACGGGTTGGGTTAGA...................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 |
| ...............................................................................................................................................................................................................................TGCGAAATCATGTGCGACAAGATGAactc | 29 | actc | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - |
| .......................................................................................................................GTATAGCCTCGAGGAGCTGAAAggg.......................................................................................................... | 25 | ggg | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - |
| ............................................................................................................................................................................TACTGACTGCTGGCCTCTGTGGATC..................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - |
| ......................................................................................................TGGATTCTGGCTTGGGAGTATAGCCTC......................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - |
| .......................................................................................................................................................................................................................TGACGAAATGCGAAATCATGTGCGACA........ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - |
| ...................................................................................................ATGTGGATTCTGGCTTGGGAGTATA.............................................................................................................................. | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - |
| ................................................................................................TAGATGTGGATTCTGGCTTGGGAG.................................................................................................................................. | 24 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - |
| AAATGAGATTTATAAATAAAGGCTAATTTTTTATCAGTCCAAAACAACCCCAGAGGCTGAGACTCTTGCTGTCTGGCAGGTTATCACGGGTTGGGTTAGATGTGGATTCTGGCTTGGGAGTATAGCCTCGAGGAGCTGAAATGGGTATGGGGCACCAGATGCCCAGGGGTCCTACTGACTGCTGGCCTCTGTGGATCCAGGTCAGCACCTCGGCATGACGAAATGCGAAATCATGTGCGACAAGATGACC ......................................................................................................(((((((.(((.(((.(((.(((.....((((.((....(((((((((....)))..)))))).)).)))).)))))).)))))).....)))))))................................................... ...........................................................................................92..........................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) |
|---|