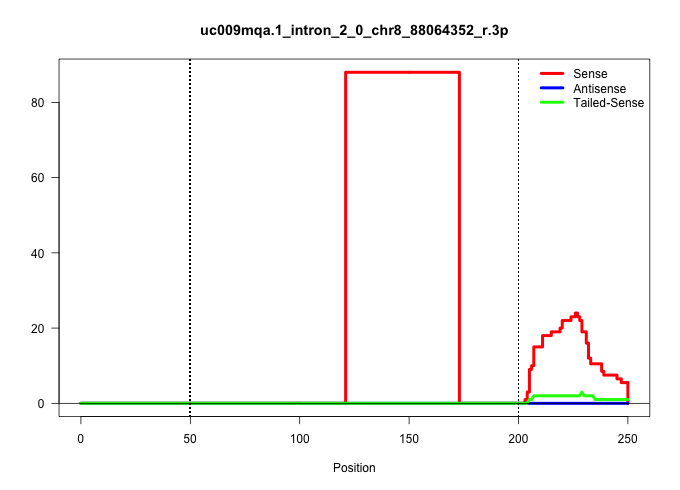
| Gene: Dnaja2 | ID: uc009mqa.1_intron_2_0_chr8_88064352_r.3p | SPECIES: mm9 |
 |
|
(2) PIWI.ip |
(1) PIWI.mut |
(12) TESTES |
| ATTGGGATCATGTAATAGTTTCTGGCATATTCCTTGAGAGTAGCTAGCTCTGCTAATAAAAAGCTTTGAAATTAGGGACAGTGGCTTTTTGGCTCTGGTAGTGAATGGTAAAGGTAAGATTCTGAAATTGACTTAAGGAATGCTCAGCAGGGTAATGTGTTTTTGTATTTTAATATATTTTGTATATACTTTGTCTATAGGTGTTCCAGAGAGATGGGAATGATTTGCATATGACATATAAGATAGGACT |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR363958(GSM822760) Adult-WTSmall RNA Miwi IP. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................CTGAAATTGACTTAAGGAATGCTCAGCAGGGTAATGTGTTTTTGTATTTTAA............................................................................. | 52 | 1 | 88.00 | 88.00 | 88.00 | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................CCAGAGAGATGGGAATGATTTGCATA................... | 26 | 1 | 3.00 | 3.00 | - | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................TGATTTGCATATGACATATAAGATAGGACT | 30 | 1 | 2.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - |
| .............................................................................................................................................................................................................CCAGAGAGATGGGAATGATTTGCA..................... | 24 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................AGAGAGATGGGAATGATTTGCATATG................. | 26 | 1 | 2.00 | 2.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................AGATGGGAATGATTTGCATATGACATA............ | 27 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - |
| ................................................................................................................................................................................................................................TTGCATATGACATATAAGATAGGACT | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................AGATGGGAATGATTTGCATATGACATAT........... | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................ATGATTTGCATATGACATATAAGATAGGACT | 31 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................................TGCATATGACATATAAGATAGG... | 22 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................TTCCAGAGAGATGGGAATGATTTGCATAT.................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .............................................................................................................................................................................................................CCAGAGAGATGGGAATGATTTGCATAT.................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................CCAGAGAGATGGGAATGATTTGCAa.................... | 25 | a | 1.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................TCCAGAGAGATGGGAATGATTTGCATAT.................. | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................GGGAATGATTTGCATATGACATATAAGATA..... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...............................................................................................................................................................................................................AGAGAGATGGGAATGATTTGCATATGAt............... | 28 | t | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................AGAGAGATGGGAATGATTTGCATAT.................. | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................CAGAGAGATGGGAATGATTTGCA..................... | 23 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................TCCAGAGAGATGGGAATGATTTGC...................... | 24 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................AGAGAGATGGGAATGATTTG....................... | 20 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................................TATGACATATAAGATAGGACTtgta | 25 | tgta | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..................................................................................................................................................................................................................................GCATATGACATATAAGATAGGACT | 24 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................AGAGAGATGGGAATGATT......................... | 18 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................................ACATATAAGATAGGACT | 17 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | 0.50 |
| ATTGGGATCATGTAATAGTTTCTGGCATATTCCTTGAGAGTAGCTAGCTCTGCTAATAAAAAGCTTTGAAATTAGGGACAGTGGCTTTTTGGCTCTGGTAGTGAATGGTAAAGGTAAGATTCTGAAATTGACTTAAGGAATGCTCAGCAGGGTAATGTGTTTTTGTATTTTAATATATTTTGTATATACTTTGTCTATAGGTGTTCCAGAGAGATGGGAATGATTTGCATATGACATATAAGATAGGACT |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR363958(GSM822760) Adult-WTSmall RNA Miwi IP. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) |
|---|