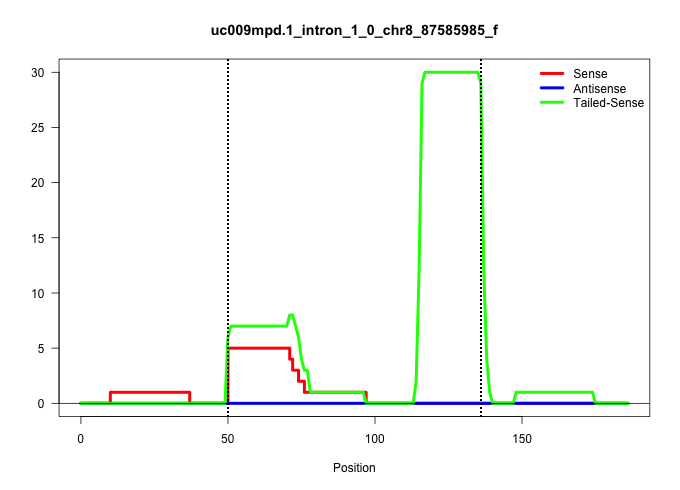
| Gene: Fbxw9 | ID: uc009mpd.1_intron_1_0_chr8_87585985_f | SPECIES: mm9 |
 |
 |
|
(9) OTHER.mut |
(1) OVARY |
(3) PIWI.ip |
(2) PIWI.mut |
(25) TESTES |
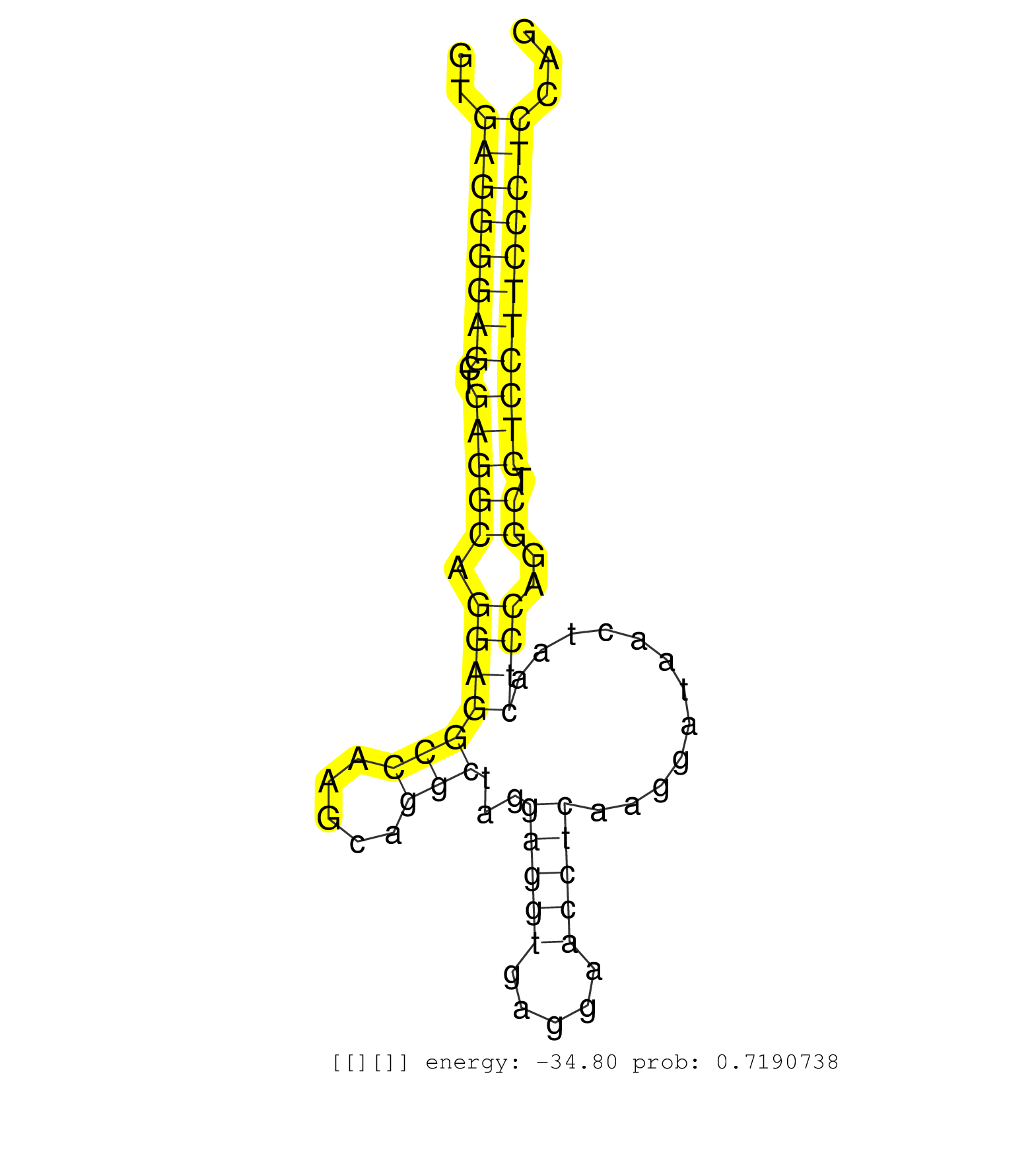
| GCCTGGCTGATGGCCACTTTGCTTCCATTGATGCGGTGTTGCTGCTCCAGGTGAGGGGAGCTGAGGCAGGAGGCCAAGCAGGCTAGGAGGTGAGGAACCTCAAGGATAACTAACTCCAGGCTCTCCTTCCCTCCAGGGTGGGGCACTGTGTCTGTCAGGCTCCCGGGATCGGAATGTCAACCTGTG ....................................................((((((((..(((((.(((((((.....)))...(((((.....)))))............))))..)).)))))))))))..................................................... ..................................................51...................................................................................136................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT1() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesWT4() Testes Data. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM475281(GSM475281) total RNA. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTGAGGGGAGCTGAGGCAGGAG.................................................................................................................. | 22 | 1 | 11.00 | 11.00 | 6.00 | 1.00 | 1.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................CAGGCTCTCCTTCCCTCCAGt................................................. | 21 | t | 10.00 | 0.00 | - | - | 2.00 | - | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGGGGAGCTGAGGCAGGAGt................................................................................................................. | 23 | t | 8.00 | 11.00 | 7.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................CCAGGCTCTCCTTCCCTCCAGt................................................. | 22 | t | 7.00 | 0.00 | - | - | 4.00 | 1.00 | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................CAGGCTCTCCTTCCCTCCAGa................................................. | 21 | a | 5.00 | 0.00 | - | - | - | - | - | 2.00 | - | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................CAGGCTCTCCTTCCCTCCAagt................................................ | 22 | agt | 5.00 | 0.00 | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 |
| ..................................................GTGAGGGGAGCTGAGGCAGGAGGC................................................................................................................ | 24 | 1 | 5.00 | 5.00 | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...................................................................................................................CCAGGCTCTCCTTCCCTCCAGa................................................. | 22 | a | 4.00 | 0.00 | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGGGGAGCTGAGGCAGGAGtta............................................................................................................... | 25 | tta | 4.00 | 11.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TGAGGGGAGCTGAGGCAGGAGG................................................................................................................. | 22 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGGGGAGCTGAGGCAGGAGGCCAAGCAGGCTAGG................................................................................................... | 37 | 1 | 3.00 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGGGGAGCTGAGGCAGGAGa................................................................................................................. | 23 | a | 3.00 | 11.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................CCAGGCTCTCCTTCCCTCCAGtat............................................... | 24 | tat | 2.00 | 0.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGGGGAGCTGAGGCAGGA................................................................................................................... | 21 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGGGGAGCTGAGGCAGGAGGCCAtt............................................................................................................ | 28 | tt | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - |
| ...................................................TGAGGGGAGCTGAGGCAGGAGGtaa.............................................................................................................. | 25 | taa | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................GGCCAAGCAGGCTAGGAGGTGAGGAA......................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ....................................................................................................................................................TGTCTGTCAGGCTCCCGGGATCGaaat........... | 27 | aaat | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................CAGGCTCTCCTTCCCTCCAGtt................................................ | 22 | tt | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................CCAGGCTCTCCTTCCCTCCAGtt................................................ | 23 | tt | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TGAGGGGAGCTGAGGCAGGAGat................................................................................................................ | 23 | at | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................GGCCAAGCAGGCTAGGAGGTGAGGAt......................................................................................... | 26 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGGGGAGCTGAGGCAGt.................................................................................................................... | 20 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..................................................GTGAGGGGAGCTGAGGCA...................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................TCCAGGCTCTCCTTCCCTCCAt.................................................. | 22 | t | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TGAGGGGAGCTGAGGCAGGAGGaa............................................................................................................... | 24 | aa | 1.00 | 3.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGGGGAGCTGAGGCAGGAGGCCA.............................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGGGGAGCTGAGGCAGGAGta................................................................................................................ | 24 | ta | 1.00 | 11.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................CAGGCTCTCCTTCCCTCCAGtat............................................... | 23 | tat | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................AGGCTCTCCTTCCCTCCAagt................................................ | 21 | agt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................CAGGCTCTCCTTCCCTCCAGtaaa.............................................. | 24 | taaa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................TCCAGGCTCTCCTTCCCTCCAGa................................................. | 23 | a | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGGGGAGCTGAGGCAGGAGtaa............................................................................................................... | 25 | taa | 1.00 | 11.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..........TGGCCACTTTGCTTCCATTGATGCGGT..................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGGGGAGCTGAGGCAGGAGtat............................................................................................................... | 25 | tat | 1.00 | 11.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ....................................................................................................................CAGGCTCTCCTTCCCTCCAGaag............................................... | 23 | aag | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................CAGGCTCTCCTTCCCTCCAGatgt.............................................. | 24 | atgt | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................CAGGCTCTCCTTCCCTCCAag................................................. | 21 | ag | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| GCCTGGCTGATGGCCACTTTGCTTCCATTGATGCGGTGTTGCTGCTCCAGGTGAGGGGAGCTGAGGCAGGAGGCCAAGCAGGCTAGGAGGTGAGGAACCTCAAGGATAACTAACTCCAGGCTCTCCTTCCCTCCAGGGTGGGGCACTGTGTCTGTCAGGCTCCCGGGATCGGAATGTCAACCTGTG ....................................................((((((((..(((((.(((((((.....)))...(((((.....)))))............))))..)).)))))))))))..................................................... ..................................................51...................................................................................136................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT1() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesWT4() Testes Data. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM475281(GSM475281) total RNA. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................TGCTCCAGGTGAGGcta.................................................................................................................................. | 17 | cta | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ......................................TGCTCCAGGTGAGGccat.................................................................................................................................. | 18 | ccat | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................AGGCTCCCGGGATtca................. | 16 | tca | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |