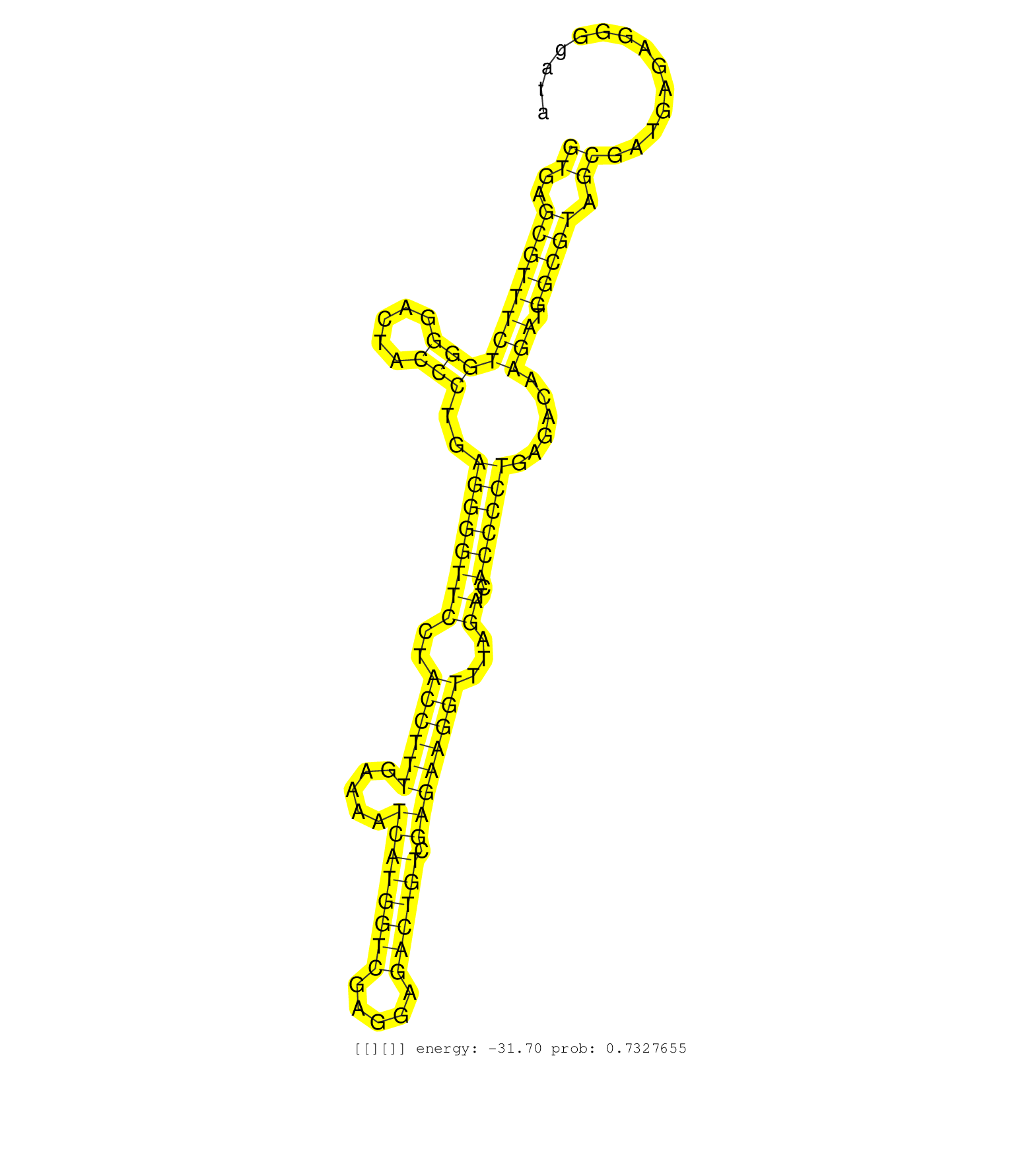

| Gene: AK157878 | ID: uc009mlj.1_intron_3_0_chr8_86460075_r.5p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(4) PIWI.ip |
(2) PIWI.mut |
(17) TESTES |
| AGCTTTAAGGAGGAGAGAGTGGCCTGGGCGGTTTAGGCGGCTGTCAGCAGGTGAGCGTTTCTGGGGACTACCCTGAGGGGTTCCTACCTTTGAAAATCATGGTCGAGGAGACTGTCGAGAAGGTTTAGATCACCCCTGAGACAAGATGGCGTAGCGATGAGAGGGGATACAGGATCGAGTCTCAGTCCGAAGGGAGTTCTGGTTCGTGGACAGACCAGGGTTTGAATCCTGCCTCTGCACTCTGTCTTTC ..................................................((..(((((((((((.....)))..((((((((..((((((.....((((((((.....)))))).))))))))...))..))))))......))).))))).))............................................................................................... ..................................................51....................................................................................................................169............................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT4() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM475281(GSM475281) total RNA. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTGAGCGTTTCTGGGGACTACCCTGAGGGGTTCCTACCTTTGAAAATCATGG.................................................................................................................................................... | 52 | 1 | 142.00 | 142.00 | 142.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................GTCGAGGAGACTGTCGAGAAGGTTTAGATCACCCCTGAGACAAGATGGCGTA................................................................................................. | 52 | 1 | 42.00 | 42.00 | - | 42.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................ATGGCGTAGCGATGAGAGGGGATACg............................................................................... | 26 | g | 8.00 | 0.00 | - | - | 8.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................TGAGACAAGATGGCGTAGCGATGAGA........................................................................................ | 26 | 1 | 4.00 | 4.00 | - | - | - | - | - | 1.00 | - | - | 2.00 | - | 1.00 | - | - | - | - | - | - |
| ......AAGGAGGAGAGAGTGGCCTGGGCGGT.......................................................................................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................AGGCGGCTGTCAGCAGGT...................................................................................................................................................................................................... | 18 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................TAGGCGGCTGTCAGCAGGTGtcct................................................................................................................................................................................................. | 24 | tcct | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........GAGGAGAGAGTGGCCTGGGC............................................................................................................................................................................................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................CGGCTGTCAGCAGGTGAGC.................................................................................................................................................................................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......AAGGAGGAGAGAGTGGCCTGG............................................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ........................................................................................TTTGAAAATCATGGTCGAGGAGACTGT....................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..........AGGAGAGAGTGGCCTGGGCa............................................................................................................................................................................................................................ | 20 | a | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGCGTTTCTGGGGACTACCCTG............................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .......................................................................................................................................................................................................TGGTTCGTGGACAGACCAGGGTTTGt......................... | 26 | t | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .GCTTTAAGGAGGAGAGAGT...................................................................................................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ........................................................................................................................AGGTTTAGATCACCCCTGAGACAAGAT....................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .................................TAGGCGGCTGTCAGCAGGc...................................................................................................................................................................................................... | 19 | c | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .............................................................................................................................................CAAGATGGCGTAGCGATGAGAGGGGA................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................TGAGACAAGATGGCGTAGCGATGAGAt....................................................................................... | 27 | t | 1.00 | 4.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................TGAAAATCATGGTCGAGGAGACTGTC...................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................GGGCGGTTTAGGCGGCTGT.............................................................................................................................................................................................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................CAAGATGGCGTAGCGATGAGAGGGGATA................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ....TTAAGGAGGAGAGAGTGGCCT................................................................................................................................................................................................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................AGACTGTCGAGAAGGattc........................................................................................................................... | 19 | attc | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .................................TAGGCGGCTGTCAGCAGGTGt.................................................................................................................................................................................................... | 21 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ....TTAAGGAGGAGAGAGTGGCCTGGG.............................................................................................................................................................................................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..........AGGAGAGAGTGGCCTGGG.............................................................................................................................................................................................................................. | 18 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............GAGAGTGGCCTGGGCGGT.......................................................................................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| AGCTTTAAGGAGGAGAGAGTGGCCTGGGCGGTTTAGGCGGCTGTCAGCAGGTGAGCGTTTCTGGGGACTACCCTGAGGGGTTCCTACCTTTGAAAATCATGGTCGAGGAGACTGTCGAGAAGGTTTAGATCACCCCTGAGACAAGATGGCGTAGCGATGAGAGGGGATACAGGATCGAGTCTCAGTCCGAAGGGAGTTCTGGTTCGTGGACAGACCAGGGTTTGAATCCTGCCTCTGCACTCTGTCTTTC ..................................................((..(((((((((((.....)))..((((((((..((((((.....((((((((.....)))))).))))))))...))..))))))......))).))))).))............................................................................................... ..................................................51....................................................................................................................169............................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT4() Testes Data. (testes) | mjTestesWT3() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM475281(GSM475281) total RNA. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) |
|---|