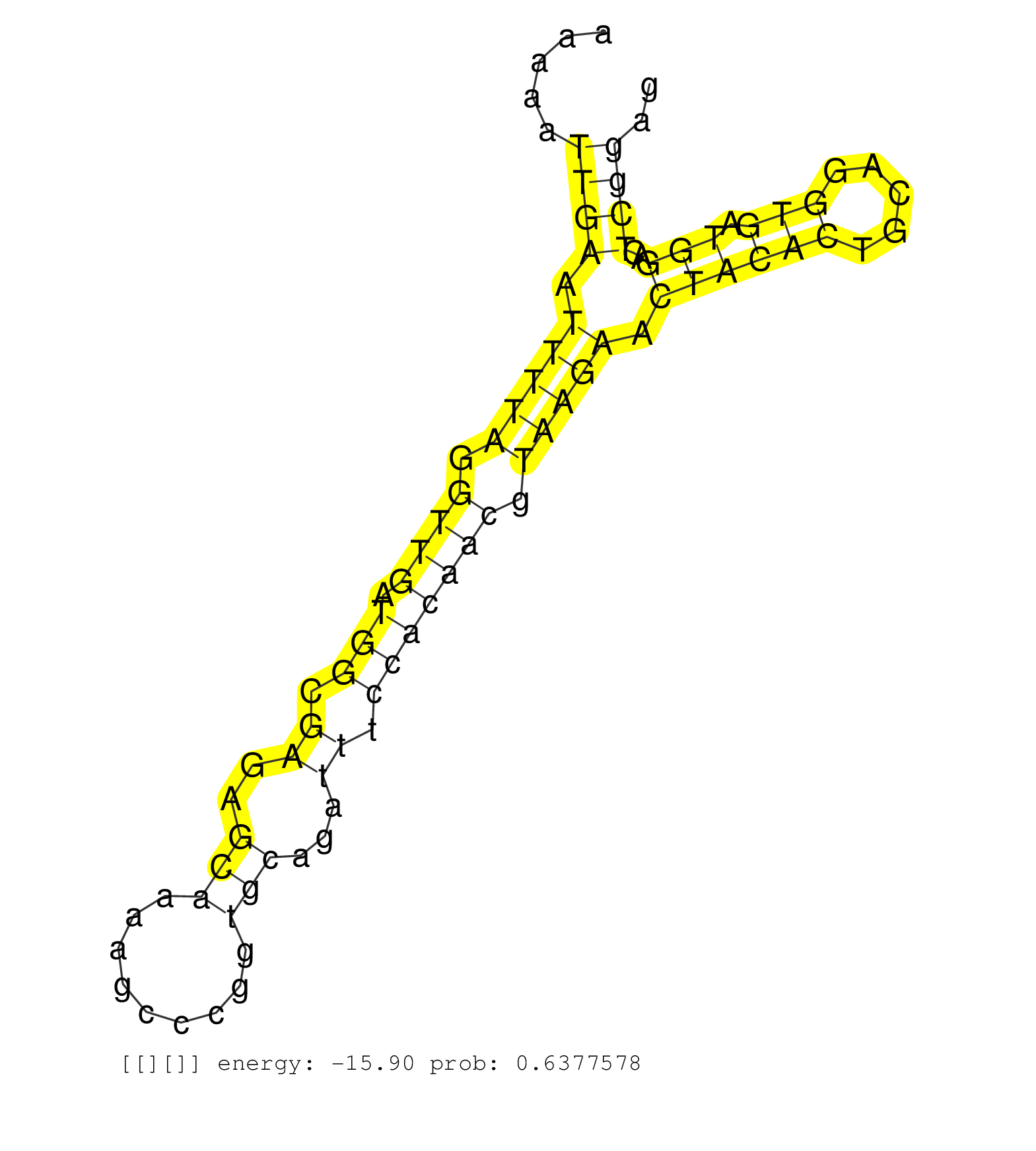

| Gene: AK134933 | ID: uc009mlf.1_intron_3_0_chr8_86414210_r.5p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(4) PIWI.ip |
(1) PIWI.mut |
(15) TESTES |
| GCAACCGGACAGGGTCCTGGAGCTGACCAGGCTGCAGGAGCGGAGGAGAGGTTAGTATCTATAAACGCCTCAAGCGTCTGGTTTTGGAGGGGGTGGGTAGGGAAGAAAAAAAAAATTGAATTTTAGGTTGATGGCGAGAGCAAAAGCCCGGTGCAGATTTCCACAACGTAAGAACTACACTGCAGGTGATGGACTCGGAGAAGTAGGGCAGCGGATCGGACTTAGATTCCTAGGCTTTGGACTTAGTGAG ...................................................................................................................((((.(((((.((((.(((.((..(((.........)))...)).))))))).))))).((((((.....))).)))..)))).................................................... ..............................................................................................................111......................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT2() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesWT3() Testes Data. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................................................................................TGGACTCGGAGAAGTAGGGCA........................................ | 21 | 1 | 7.00 | 7.00 | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................TAAGAACTACACTGCAGGTGATGGACTC...................................................... | 28 | 1 | 7.00 | 7.00 | - | - | 3.00 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - |
| ..........................................................................................................................................................................................TGATGGACTCGGAGAAGTAGGGCAGC...................................... | 26 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | 1.00 | - | - | - |
| .......................................................................................................................................................................................................................TCGGACTTAGATTCCTAGGCTTTGGACTT...... | 29 | 1 | 3.00 | 3.00 | - | - | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................TAAGAACTACACTGCAGGTGATGGACT....................................................... | 27 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................TACACTGCAGGTGATGGACTCGGAGA................................................. | 26 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TGCAGGTGATGGACTCGGAGAAGTAGG........................................... | 27 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................TTTCCACAACGTAAGAACTACACTGC................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................TAGATTCCTAGGCTTTGGACTTAGT... | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................TTGAATTTTAGGTTGATGGCGAGAGC............................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................TCGGACTTAGATTCCTAGGCTTTGGACT....... | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................TACACTGCAGGTGATGGACTCGGAG.................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................GGTGATGGACTCGGAGAAGTAGGGC......................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................TAGATTCCTAGGCTTTGGACTTAG.... | 24 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................ACTGCAGGTGATGGccct...................................................... | 18 | ccct | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ...........................................................................................................................................................................GAACTACACTGCAGGTGATGGACTCGGA................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................TTCCACAACGTAAGAACTACACTGCAGG................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ........................................................................................................................TTTTAGGTTGATGGCGAGAGCAAAAGC....................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...................................................................................TTGGAGGGGGTGGGTAGGGAA.................................................................................................................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ........................................................................................................................................................................TAAGAACTACACTGCAGGTGATGGAC........................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...................................................................................................................................................................CAACGTAAGAACTACACTGCAGGTGATG........................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...................................................................................................................TTGAATTTTAGGTTGATGGCGAGAG.............................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .............................................................................................................................................................TTTCCACAACGTAAGAACTACACTGCA.................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| GCAACCGGACAGGGTCCTGGAGCTGACCAGGCTGCAGGAGCGGAGGAGAGGTTAGTATCTATAAACGCCTCAAGCGTCTGGTTTTGGAGGGGGTGGGTAGGGAAGAAAAAAAAAATTGAATTTTAGGTTGATGGCGAGAGCAAAAGCCCGGTGCAGATTTCCACAACGTAAGAACTACACTGCAGGTGATGGACTCGGAGAAGTAGGGCAGCGGATCGGACTTAGATTCCTAGGCTTTGGACTTAGTGAG ...................................................................................................................((((.(((((.((((.(((.((..(((.........)))...)).))))))).))))).((((((.....))).)))..)))).................................................... ..............................................................................................................111......................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT4() Testes Data. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT2() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesWT3() Testes Data. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|