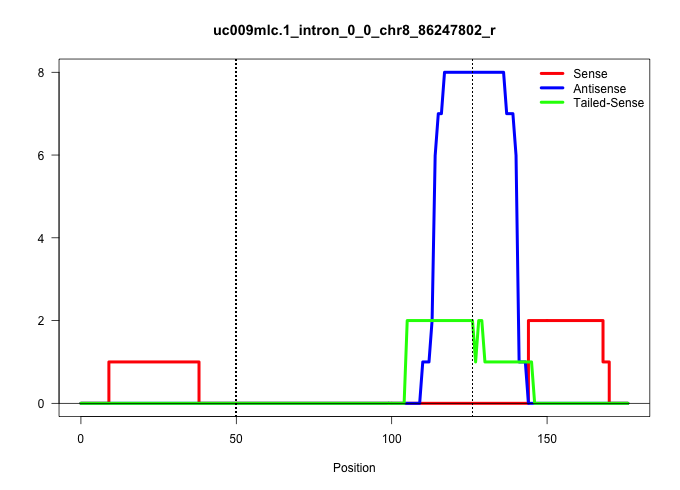
| Gene: Cd97 | ID: uc009mlc.1_intron_0_0_chr8_86247802_r | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(5) PIWI.ip |
(17) TESTES |
| ACACAGAGTTCAACTCTTCCACAACAGGCACTGGCACCAGCCAAACACGGGTAAGGGGCTGGCCTGCAGTGGGGCTGGGAGGGCAGGCCTAGGTGAGCTCAGCCTCAAAGGCCTCCTTCTCTGCAGGCTCTCAGGTCCTCAGAATCAGGGATGTGAAGGCGAGTTCCATGAAGGAC ..................................................((((((((..(((((....((((((((((.(((....))).......)))))))))).)))))..))))).))).................................................... ..................................................51.........................................................................126................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | mjTestesWT1() Testes Data. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | mjTestesWT2() Testes Data. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................CAAAGGCCTCCTTCTCTGCAG.................................................. | 21 | 1 | 6.00 | 6.00 | 4.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................CAAAGGCCTCCTTCTCTGCAGt................................................. | 22 | t | 4.00 | 6.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .........................................................................................................CAAAGGCCTCCTTCTCTGCAGtaa............................................... | 24 | taa | 2.00 | 6.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................CAAAGGCCTCCTTCTCTGCAGttgt.............................................. | 25 | ttgt | 1.00 | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ................................................................................................................................................TCAGGGATGTGAAGGCGAGTTCCATG...... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .......GTTCAACTCTTCCACAACAGGC................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ................................................................................................................................TCTCAGGTCCTCAGAAct.............................. | 18 | ct | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .........TCAACTCTTCCACAACAGGCACTGGCACC.......................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................TCAGGGATGTGAAGGCGAGTTCCA........ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ACACAGAGTTCAACTCTTCCACAACAGGCACTGGCACCAGCCAAACACGGGTAAGGGGCTGGCCTGCAGTGGGGCTGGGAGGGCAGGCCTAGGTGAGCTCAGCCTCAAAGGCCTCCTTCTCTGCAGGCTCTCAGGTCCTCAGAATCAGGGATGTGAAGGCGAGTTCCATGAAGGAC ..................................................((((((((..(((((....((((((((((.(((....))).......)))))))))).)))))..))))).))).................................................... ..................................................51.........................................................................126................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | mjTestesWT1() Testes Data. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | mjTestesWT2() Testes Data. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................CCTTCTCTGCAGGCTCTCAGGTCCTCA................................... | 27 | 1 | 3.00 | 3.00 | - | - | - | - | 1.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................CCTTCTCTGCAGGCTCTCAGGTCC...................................... | 24 | 1 | 2.00 | 2.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................CCTTCTCTGCAGGCTCTCAGGTCCTCAtt................................... | 29 | tt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...................................................................................................................CTTCTCTGCAGGCTCTCAGGTCCTCA................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................TCTCTGCAGGCTCTCAGGTCCTCAGAA................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..........................................................................................................................GCTCTCAGGTCCTCAtgag................................... | 19 | tgag | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................TCCTTCTCTGCAGGCTCTCAGGTCCTCA................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................CAGCCTCAAAGGCCTCCTTCTCTGCAggg................................................... | 29 | ggg | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................CCTTCTCTGCAGGCTCTCAGGTCCTC.................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................GCCTCCTTCTCTGCAGGCTCTCAGGTC....................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |