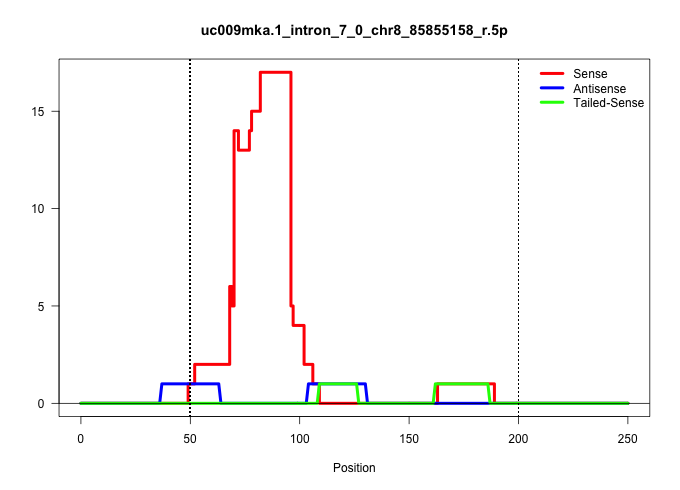
| Gene: Elmod2 | ID: uc009mka.1_intron_7_0_chr8_85855158_r.5p | SPECIES: mm9 |
 |
|
(6) OTHER.mut |
(1) PIWI.mut |
(13) TESTES |
| AGCTGGGGAAACCGAGGCAGTGCCAGCTTCCCCCTAGTTCTTCCGCTCCTGTGAGGTGGCTGCTGTTTTATTTTTCTCTCACTCTGAGTTGCTGGCGTGCGGCTGCGCAAGCCTCGCTGGCGTCCCGCGGATAACCCGCAGGGTGCGGGGGCCGTCGGGGCTTTGTGCAGCGTTCCCGCGCGCGAGAGGGGCGGGACGGCGCGGTGCCCACGGGGCTCGCACTCGGTTTCCCGAGGCCCTGGGGCTCGGC |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT3() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | mjTestesWT2() Testes Data. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................TTTTTCTCTCACTCTGAGTTGCTGGC.......................................................................................................................................................... | 26 | 1 | 8.00 | 8.00 | 6.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ....................................................................TATTTTTCTCTCACTCTGAGTTGCTGGC.......................................................................................................................................................... | 28 | 1 | 4.00 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................AGCCTCGCTGGCGTtgat........................................................................................................................... | 18 | tgat | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ....................................................GAGGTGGCTGCTGTTTTATT.................................................................................................................................................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................TTGTGCAGCGTTCCCGCGCGCGAGt............................................................... | 25 | t | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .................................................TGTGAGGTGGCTGCTGTTTT..................................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..............................................................................TCACTCTGAGTTGCTGGCGTGCGG.................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..................................................................................TCTGAGTTGCTGGCGTGCGGCTGC................................................................................................................................................ | 24 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................TGTGCAGCGTTCCCGCGCGCGAGAGG............................................................. | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................TCTGAGTTGCTGGCGTGCGGCTGCGCA............................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .............................................................................CTCACTCTGAGTTGCTGGCGTGCGG.................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ......................................................................TTTTTCTCTCACTCTGAGTTGCTGGCG......................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| AGCTGGGGAAACCGAGGCAGTGCCAGCTTCCCCCTAGTTCTTCCGCTCCTGTGAGGTGGCTGCTGTTTTATTTTTCTCTCACTCTGAGTTGCTGGCGTGCGGCTGCGCAAGCCTCGCTGGCGTCCCGCGGATAACCCGCAGGGTGCGGGGGCCGTCGGGGCTTTGTGCAGCGTTCCCGCGCGCGAGAGGGGCGGGACGGCGCGGTGCCCACGGGGCTCGCACTCGGTTTCCCGAGGCCCTGGGGCTCGGC |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT3() Testes Data. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | mjTestesWT2() Testes Data. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................TTCTTCCGCTCCTGTGAGGTGGCTGCT.......................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................GCGCAAGCCTCGCTGGCGTCCCGCGGA....................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |