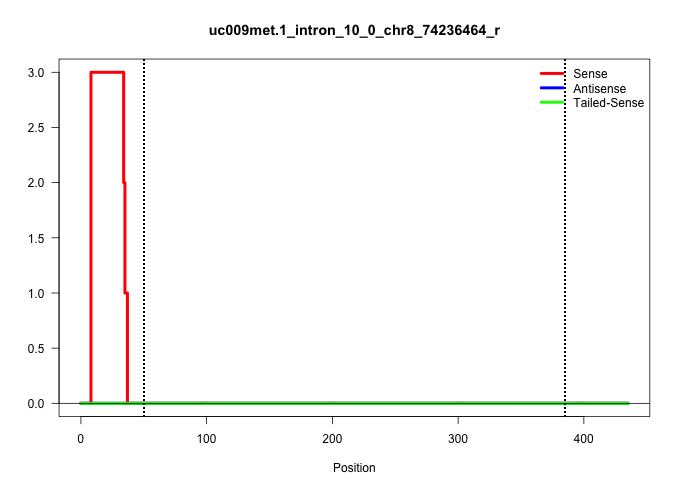
| Gene: Fcho1 | ID: uc009met.1_intron_10_0_chr8_74236464_r | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(2) PIWI.ip |
(7) TESTES |
| GACACTTCTGGGAAGCCACAGAGACCTCGTTCGGCCCCACGCACTGGCAGGTGGGTCTCACCGGGACATGATGGAGCAGGCAGATGGGCCCCAAGTCCTTGCCTCTAGCTCCCTCACCCTGGCCTAAGGTCCAGCTATTGGACACTTATTAAGTGCCAGCTGTGTACTTGTGCTGTGCTGGCTCTGGGAAGCCTGTAGACAAGGGTCAAAATTCTGGATGCGCGGATGAGTGGGATGTGGCCATGCTGCATAGGCAGGGTCCTGGGGGAAGATGTAACTACTAAACCCGAAAGGCCTAGGATTTGCGGGTGAAGTCACGCGCTGGGAACAGGAACAGCCTGTGGGAAGGTAGCAAGCTCGAACTCGCTCCTGTCGCTGTTCCCAGTTGCGCAGAGAAGCCCCTGGCCTCGGAGGAGCCACTATCCAAGAGCCTCT ..............................................................................................................................................................................................................................................................................................((....))..........(((.(((....))).))).((((((((..((((...(((((..(((.....)))....)))))..))))..)))))))).................................................... .............................................................................................................................................................................................................................................................................................286................................................................................................385................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT2() Testes Data. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT3() Testes Data. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ........TGGGAAGCCACAGAGACCTCGTTCGG................................................................................................................................................................................................................................................................................................................................................................................................................. | 26 | 1 | 2.00 | 2.00 | 1.00 | - | - | 1.00 | - | - | - |
| ..................................................................................................................................................................................................................................................................................................................................TGGGAACAGGAACAGCCTGTGG........................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - |
| ........TGGGAAGCCACAGAGACCTCGTTCGGC................................................................................................................................................................................................................................................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - |
| ........TGGGAAGCCACAGAGACCTCGTTCGGCCC.............................................................................................................................................................................................................................................................................................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - |
| ............................................................................................................................................................................CTGTGCTGGCTCTGGGA...................................................................................................................................................................................................................................................... | 17 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | 0.33 |
| GACACTTCTGGGAAGCCACAGAGACCTCGTTCGGCCCCACGCACTGGCAGGTGGGTCTCACCGGGACATGATGGAGCAGGCAGATGGGCCCCAAGTCCTTGCCTCTAGCTCCCTCACCCTGGCCTAAGGTCCAGCTATTGGACACTTATTAAGTGCCAGCTGTGTACTTGTGCTGTGCTGGCTCTGGGAAGCCTGTAGACAAGGGTCAAAATTCTGGATGCGCGGATGAGTGGGATGTGGCCATGCTGCATAGGCAGGGTCCTGGGGGAAGATGTAACTACTAAACCCGAAAGGCCTAGGATTTGCGGGTGAAGTCACGCGCTGGGAACAGGAACAGCCTGTGGGAAGGTAGCAAGCTCGAACTCGCTCCTGTCGCTGTTCCCAGTTGCGCAGAGAAGCCCCTGGCCTCGGAGGAGCCACTATCCAAGAGCCTCT ..............................................................................................................................................................................................................................................................................................((....))..........(((.(((....))).))).((((((((..((((...(((((..(((.....)))....)))))..))))..)))))))).................................................... .............................................................................................................................................................................................................................................................................................286................................................................................................385................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | mjTestesWT2() Testes Data. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT3() Testes Data. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................TCACCGGGACATGAcctt............................................................................................................................................................................................................................................................................................................................................................................ | 18 | cctt | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - |
| ..................................................................................GGCCCCAAGTCCTTGatgg.............................................................................................................................................................................................................................................................................................................................................. | 19 | atgg | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - |