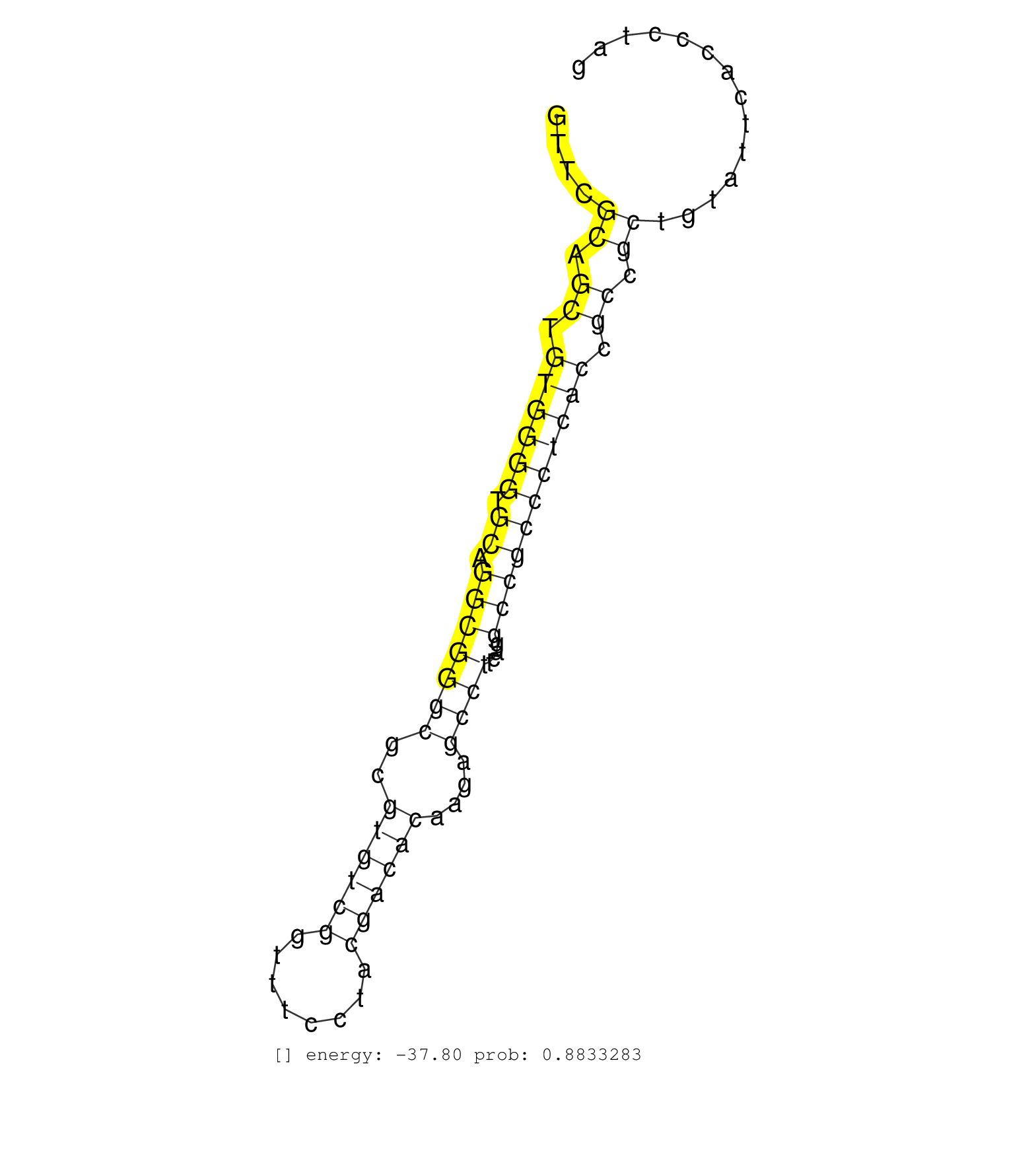
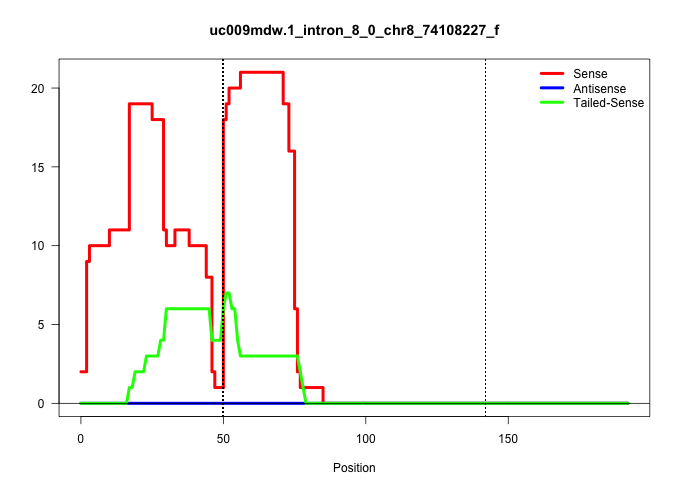
| Gene: Slc27a1 | ID: uc009mdw.1_intron_8_0_chr8_74108227_f | SPECIES: mm9 |
 |
 |
|
(6) OTHER.mut |
(4) PIWI.ip |
(2) PIWI.mut |
(24) TESTES |
| TCTACGGCGCTACCGAGTGCAACTGCAGCATTGCCAACATGGACGGCAAGGTTCGCAGCTGTGGGGTGCAGGCGGGCGCGTGTCGGTTTCCTACGACACAAGAGCCTTCAGGCCGCCCTCACCGCCGCTGTATTCACCCTAGGTCGGCTCCTGCGGCTTCAACAGCCGTATCCTCACGCATGTGTACCCCAT ......................................................((.((.((((((.((.(((((((..((((((........))))))....))))....))))))))))).)).))................................................................ ..................................................51.........................................................................................142................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT4() Testes Data. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | mjTestesWT1() Testes Data. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | mjTestesWT2() Testes Data. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTTCGCAGCTGTGGGGTGCAGGCGG..................................................................................................................... | 25 | 1 | 16.00 | 16.00 | 3.00 | - | 3.00 | - | - | 1.00 | - | - | - | 2.00 | 1.00 | 3.00 | 1.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - |
| ..................................................GTTCGCAGCTGTGGGGTGCAGGCG...................................................................................................................... | 24 | 1 | 6.00 | 6.00 | - | - | 2.00 | - | - | - | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTTCGCAGCTGTGGGGTGCAGGC....................................................................................................................... | 23 | 1 | 6.00 | 6.00 | 2.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..TACGGCGCTACCGAGTGCAACTGCAGC................................................................................................................................................................... | 27 | 1 | 6.00 | 6.00 | - | 3.00 | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................TGCAACTGCAGCATTGCCAACATGGACGG.................................................................................................................................................. | 29 | 1 | 6.00 | 6.00 | - | 3.00 | - | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTTCGCAGCTGTGGGGTGCAGGCGGa.................................................................................................................... | 26 | a | 4.00 | 16.00 | - | - | - | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTTCGCAGCTGTGGGGTGCAGGCGGG.................................................................................................................... | 26 | 1 | 3.00 | 3.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...................................................TTCGCAGCTGTGGGGTGCAGGCGGGCGCGT............................................................................................................... | 30 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTTCGCAGCTGTGGGGTGCAG......................................................................................................................... | 21 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .................TGCAACTGCAGCATTGCCAACATGGAC.................................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...ACGGCGCTACCGAGTGCAACTGCAGCA.................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................TGCAACTGCAGCATTGCCAACATGGACtg.................................................................................................................................................. | 29 | tg | 1.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................CATTGCCAACATGGACGGCAAGGTcgg......................................................................................................................................... | 27 | cgg | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TTCGCAGCTGTGGGGTGCAGGCGGGC................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ..................................................GTTCGCAGCTGTGGGGTGCAGGCGGGa................................................................................................................... | 27 | a | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .......................TGCAGCATTGCCAACATGGACGGCAAGGTc........................................................................................................................................... | 30 | c | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..................................................GTTCGCAGCTGTGGGGTGCAGG........................................................................................................................ | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TTCGCAGCTGTGGGGTGCAGGCGGGCGCGa............................................................................................................... | 30 | a | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTTCGCAGCTGTGGGGTGCAGGCGGGCt.................................................................................................................. | 28 | t | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TTCGCAGCTGTGGGGTGCAGGCGGGCGt................................................................................................................. | 28 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..TACGGCGCTACCGAGTGCAA.......................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................TTGCCAACATGGACGGCAAGGTcgg......................................................................................................................................... | 25 | cgg | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................TCGCAGCTGTGGGGTGCAGGCGG..................................................................................................................... | 23 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................CCAACATGGACGGCAAG.............................................................................................................................................. | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ........................................................AGCTGTGGGGTGCAGGCGGGCGCGTGTCG........................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...................................................TTCGCAGCTGTGGGGTGCAGGCGGG.................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTTCGCAGCTGTGGGGTGCAGGCGGGaa.................................................................................................................. | 28 | aa | 1.00 | 3.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTTCGCAGCTGTGGGGTGCAGGCGa..................................................................................................................... | 25 | a | 1.00 | 6.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................TTGCCAACATGGACGGCAAGGTcggc........................................................................................................................................ | 26 | cggc | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................CAACTGCAGCATTGCCAACATGGACag.................................................................................................................................................. | 27 | ag | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................CTGCAGCATTGCCAACATGGACGGC................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTTCGCAGCTGTGGGGTGCAGGCGGGC................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........TACCGAGTGCAACTGCAGCATTGCCAAC.......................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TCTACGGCGCTACCGAGTGCAACTGCAGCATTGCCAACATGGACGGCAAGGTTCGCAGCTGTGGGGTGCAGGCGGGCGCGTGTCGGTTTCCTACGACACAAGAGCCTTCAGGCCGCCCTCACCGCCGCTGTATTCACCCTAGGTCGGCTCCTGCGGCTTCAACAGCCGTATCCTCACGCATGTGTACCCCAT ......................................................((.((.((((((.((.(((((((..((((((........))))))....))))....))))))))))).)).))................................................................ ..................................................51.........................................................................................142................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT4() Testes Data. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | mjTestesWT1() Testes Data. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | mjTestesWT2() Testes Data. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CGGCGCTACCGgtac................................................................................................................................................................................. | 15 | gtac | 2.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |