
| Gene: Plvap | ID: uc009mdp.1_intron_0_0_chr8_74022305_r.5p | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(1) OVARY |
(5) PIWI.ip |
(2) PIWI.mut |
(17) TESTES |
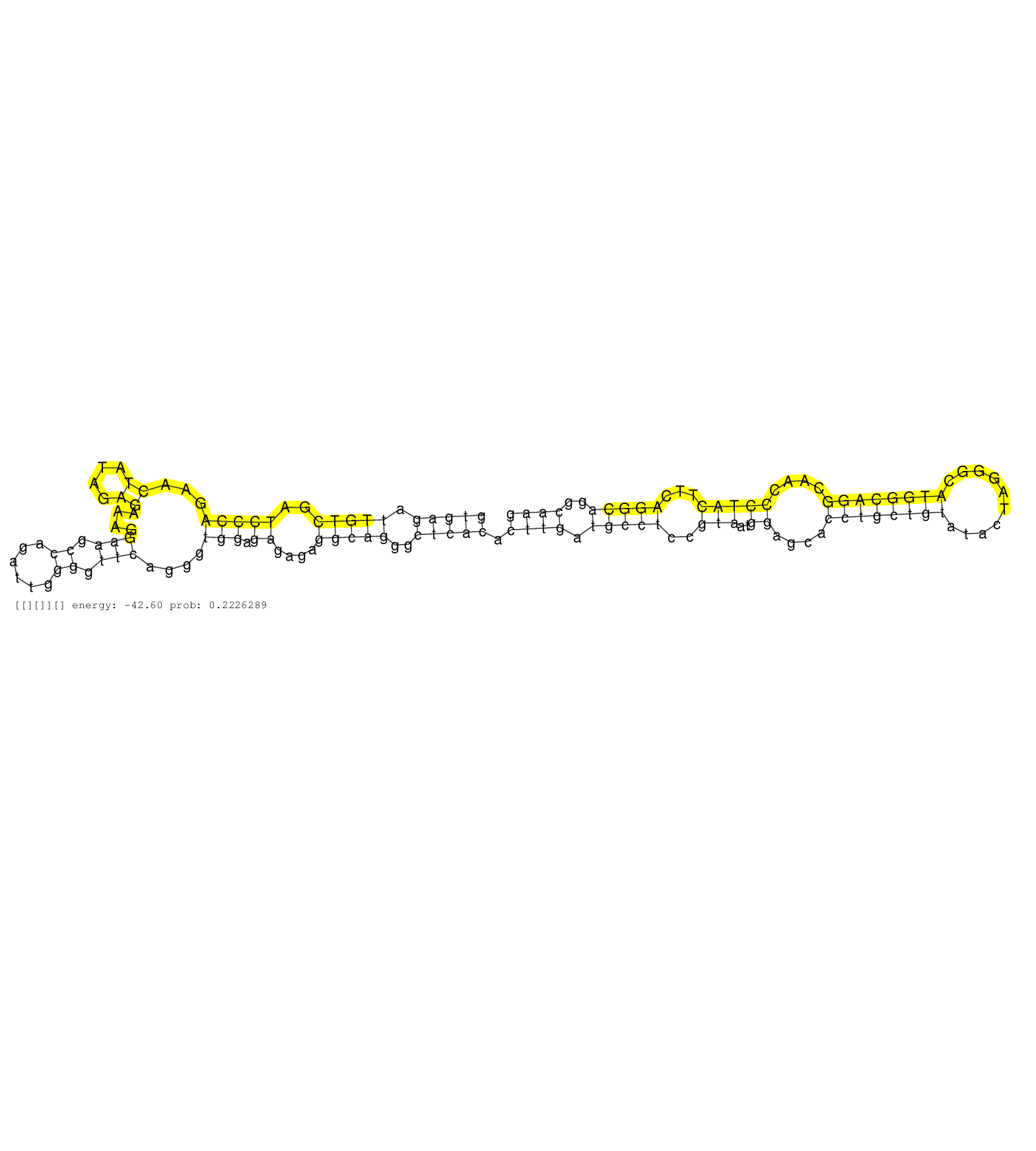
| ATCCTGGAGTCTCAGCGGCTCCCTGTAGTCAACCCTGCTGCCCAACCCAGGTGAGATTGTCGATCCCAGAACTATAGAGAAAGGAAGCCAGATTGGGGTTCAGGGTGGAGAGAGAGGCAGGGCTCACACTTGATGCCTCCGTAATGGAGCACCTGCTGTATACTAGGGCATGGCAGGCAACCCTACTTCAGGCAGGCAAGCCAGGGCTTTAGAGGGAGTACTTGTGAGCTGGAGGATGGAGAGGAGGCAG ..................................................(((((.(((((..(((((...((....))....(((.((......)).)))....))).))....)))))..))))).((((.(((((..(((..((....((((((((..........))))))))....)))))...)))))..)))).................................................. ..................................................51...................................................................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | mjTestesWT3() Testes Data. (testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................TGCTGCCCAACCCAGGTGAGATTGTC............................................................................................................................................................................................. | 26 | 1 | 3.00 | 3.00 | - | - | - | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .........................................................TGTCGATCCCAGAACTATAGAGAAAGG...................................................................................................................................................................... | 27 | 1 | 3.00 | 3.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - |
| .........................................................TGTCGATCCCAGAACTATAGAGAAAGGA..................................................................................................................................................................... | 28 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................TAGAGAAAGGAAGCCAGATTGGGGTTC..................................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................TGTCGATCCCAGAACTATAGAGAAAGGAA.................................................................................................................................................................... | 29 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .................................................................................................................................................................................................................TAGAGGGAGTACTTGTGAGCTGGAGGA.............. | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .............................................................GATCCCAGAACTATAGAGAAAGG...................................................................................................................................................................... | 23 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................TAGAGAAAGGAAGCCAGATTGGGGTT...................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..........................................................................TAGAGAAAGGAAGCCAGATTGGGGT....................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..........................................................GTCGATCCCAGAACTATAGAGAAAGGA..................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................GATCCCAGAACTATAGAGAAAGGAAGCC................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................TTAGAGGGAGTACTTGTGAGCTGG.................. | 24 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................TATAGAGAAAGGAAGCCAGATTGGGGT....................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGATTGTCGATCCCAGAACTATAGAGA.......................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .........................................................TGTCGATCCCAGAACTATAGAGAAAG....................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................TACTTCAGGCAGGCAAGCCAGGGCTTa........................................ | 27 | a | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..........................................................................TAGAGAAAGGAAGCCAGATTGGGGct...................................................................................................................................................... | 26 | ct | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...............................................................TCCCAGAACTATAGAGAAAGGAAGCCA................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ...................................................................................................................................................................TAGGGCATGGCAGGCAACCCTACTTCAGGC......................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................CTGTATACTAGGGCATGGCAGGCAAC..................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................TGTAGTCAACCCTGCTGCCCAACCa.......................................................................................................................................................................................................... | 25 | a | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................CCCAGAACTATAGAGAAAGGAAGCCAG............................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................TAGAGGGAGTACTTGTGAGCTGGA................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ATCCTGGAGTCTCAGCGGCTCCCTGTAGTCAACCCTGCTGCCCAACCCAGGTGAGATTGTCGATCCCAGAACTATAGAGAAAGGAAGCCAGATTGGGGTTCAGGGTGGAGAGAGAGGCAGGGCTCACACTTGATGCCTCCGTAATGGAGCACCTGCTGTATACTAGGGCATGGCAGGCAACCCTACTTCAGGCAGGCAAGCCAGGGCTTTAGAGGGAGTACTTGTGAGCTGGAGGATGGAGAGGAGGCAG ..................................................(((((.(((((..(((((...((....))....(((.((......)).)))....))).))....)))))..))))).((((.(((((..(((..((....((((((((..........))))))))....)))))...)))))..)))).................................................. ..................................................51...................................................................................................................................................200................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR014234(GSM319958) Ovary total. (ovary) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | mjTestesWT3() Testes Data. (testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................................................................CATGGCAGGCAACCCTACTTCAGGCAGG...................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .................................................GGTGAGATTGTCGATCCCAGAACTATA.............................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................GTACTTGTGAGCTGGAGGATGGAGAc........ | 26 | c | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................GGCAGGGCTCACACTTGATGCCTCCGTAA.......................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................TGGCAGGCAACCCTACTTCAGGCA........................................................ | 24 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................TAATGGAGCACCTGCTGTATACTA..................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |