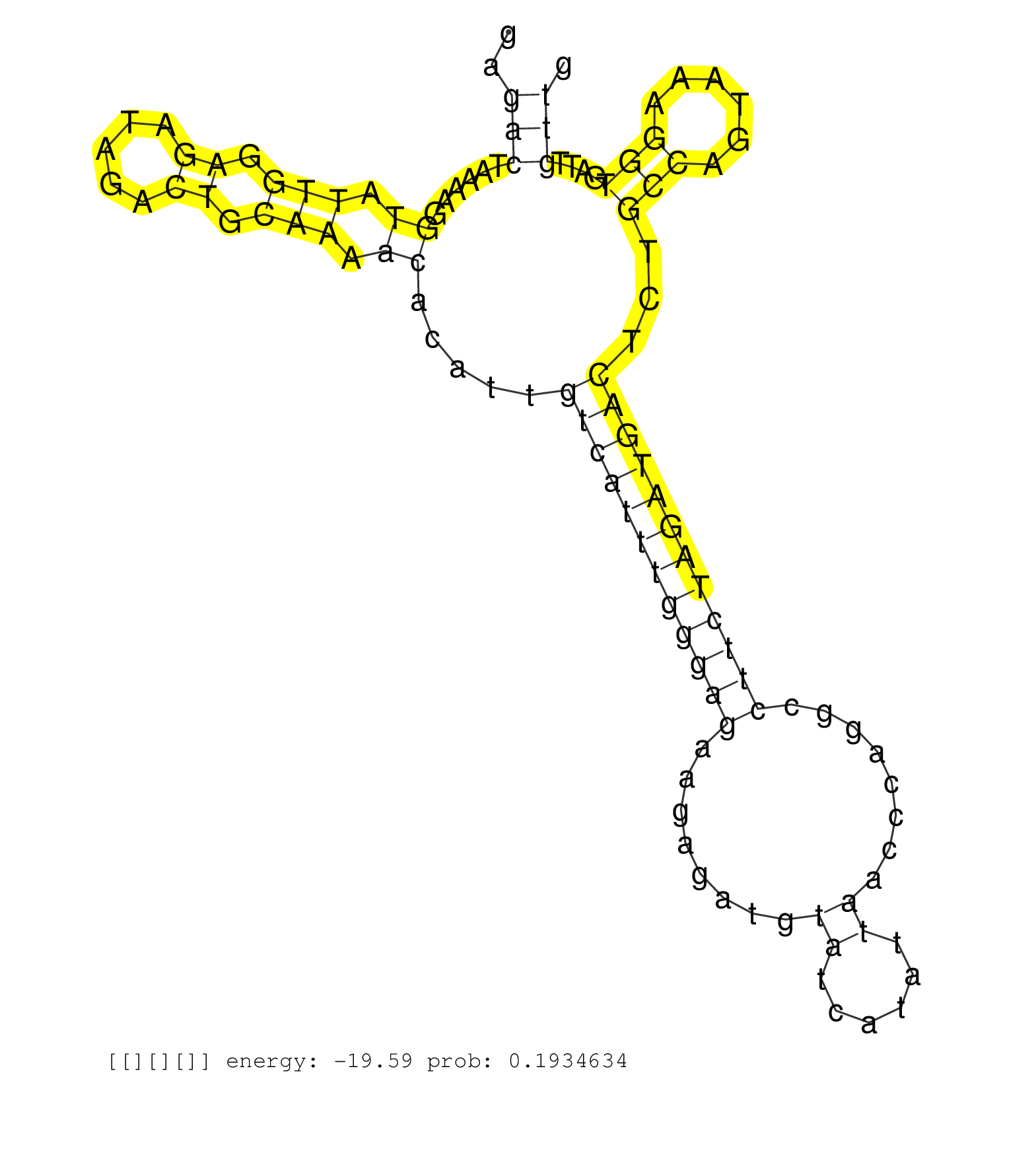

| Gene: Gtpbp3 | ID: uc009mdn.1_intron_2_0_chr8_74013414_f.5p | SPECIES: mm9 |
 |
 |
|
(1) OTHER.mut |
(5) PIWI.ip |
(1) PIWI.mut |
(17) TESTES |
| TCACGTACACGGAGGTCCTGCGGTGGTCAGCGGTGTCTTGCAGGCATTGGGTGAGGCTCTGTTGAGTGGCACTGTTCCTGGGGGAGACTAAAAGGTATTGGAGATAGACTGCAAAACACATTGTCATTTGGGAGAAGAGATGTATCATATTAACCCAGGCCTTCTAGATGACTCTGCCAGTAAAGGTGATTGTTGCCAAATGTGAACCCGAGTGGTGAAAGGTGAGAATTGATTCCTGCAGGCTGTCTTT .....................................................................................(((......((.(((.((.....)).))).)).....((((((((((((........((......))........))))))))))))...(((......)))....)))........................................................ ...................................................................................84.............................................................................................................195..................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................................TAGATGACTCTGCCAGTAAAGGTGATT........................................................... | 27 | 1 | 6.00 | 6.00 | 2.00 | - | - | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................TAAAAGGTATTGGAGATAGACTGCAAA....................................................................................................................................... | 27 | 1 | 5.00 | 5.00 | - | 3.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .................................................................................................................................................................TTCTAGATGACTCTGCCAGTAAAGGTGATT........................................................... | 30 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................TTCTAGATGACTCTGCCAGTAAAGGTGt............................................................. | 28 | t | 2.00 | 2.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................TTCTAGATGACTCTGCCAGTAAAGGTG.............................................................. | 27 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................TCTAGATGACTCTGCCAGTAAAGGTGATT........................................................... | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TGAGGCTCTGTTGAGTGGCACTGTTCt............................................................................................................................................................................ | 27 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .................................................................................................TTGGAGATAGACTGCAAAACACATTGT.............................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .................................................................................................TTGGAGATAGACTGCAAAACACATTGTC............................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..............................................................TGAGTGGCACTGTTCCTGGGGGAGAC.................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................TGAGTGGCACTGTTCCTGGGGGAGACT................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ....................................................................................................................................................................................TAAAGGTGATTGTTGCCAAATGTGAACC.......................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................TTCTAGATGACTCTGCCAGTAAAagtg.............................................................. | 27 | agtg | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....TACACGGAGGTCCTGCGGTGGTCAGt........................................................................................................................................................................................................................... | 26 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .....TACACGGAGGTCCTGCGGTGGTCAGC........................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ........................................................................................TAAAAGGTATTGGAGATAGACTGCA......................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................TAGATGACTCTGCCAGTAAAGGTGATTG.......................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...........................................................TGTTGAGTGGCACTGTTCCTGGGGG...................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ........................................................................................TAAAAGGTATTGGAGATAGACTGCAAAA...................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................TTGGGAGAAGAGATGTATCATATTAACC............................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................TTAACCCAGGCCTTCTAGATGACTCTGC......................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................TTCTAGATGACTCTGCCAGTAAAGG................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................TGTTGAGTGGCACTGTTCCTGGGGGA..................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................GTCAGCGGTGTCTTGCAGGCATTGGG....................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...................................................................................................................................................TATTAACCCAGGCCTTCTAGATGACTC............................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ................................................................................................................................................TCATATTAACCCAGGCCTTCTAGATGA............................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| TCACGTACACGGAGGTCCTGCGGTGGTCAGCGGTGTCTTGCAGGCATTGGGTGAGGCTCTGTTGAGTGGCACTGTTCCTGGGGGAGACTAAAAGGTATTGGAGATAGACTGCAAAACACATTGTCATTTGGGAGAAGAGATGTATCATATTAACCCAGGCCTTCTAGATGACTCTGCCAGTAAAGGTGATTGTTGCCAAATGTGAACCCGAGTGGTGAAAGGTGAGAATTGATTCCTGCAGGCTGTCTTT .....................................................................................(((......((.(((.((.....)).))).)).....((((((((((((........((......))........))))))))))))...(((......)))....)))........................................................ ...................................................................................84.............................................................................................................195..................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................................................................................................GATTCCTGCAGGCTcaga...... | 18 | caga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |