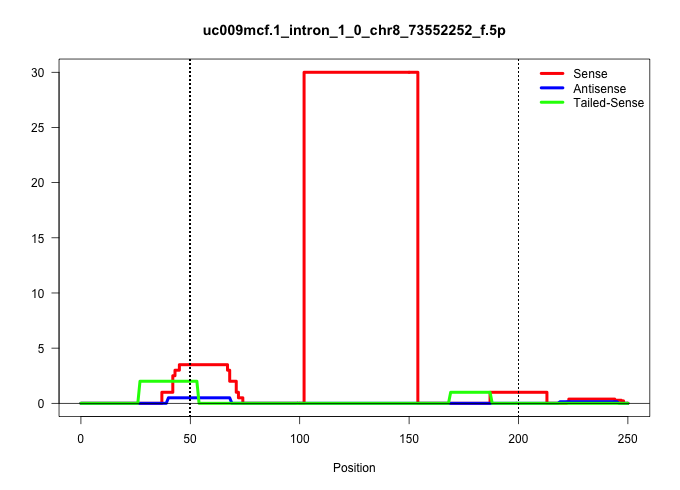
| Gene: AK029875 | ID: uc009mcf.1_intron_1_0_chr8_73552252_f.5p | SPECIES: mm9 |
 |
|
(3) OTHER.mut |
(2) PIWI.ip |
(1) PIWI.mut |
(12) TESTES |
| TTATGGTTGAGGTATGCACCCAGCCCATTGATGCCATTACTCTACTGAAGGTAAAGAATGATTCTAGCAATTCCACCCAACACACAGACAAAAGGTGCCATAGACTAACCTCTATTGAGTCTCTGATCTGTAGGGAATGGGTTTCTGATTGCAGGTGAGTAAGGGTCAGCGAGAATTGACAGACAGATGCAGACACAAGGGAGCGCTGAATCTGAATGTATTTTGTCCAAACGAACACCAGACTTTTAAT |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR363960(GSM822762) Adult Library#1Small RNA Miwi IPDuplexed run:. (testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | mjTestesWT4() Testes Data. (testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................GACTAACCTCTATTGAGTCTCTGATCTGTAGGGAATGGGTTTCTGATTGCAG................................................................................................ | 52 | 1 | 30.00 | 30.00 | 30.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...........................TTGATGCCATTACTCTACTGAAGatgt.................................................................................................................................................................................................... | 27 | atgt | 2.00 | 0.00 | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - |
| ...........................................................................................................................................................................................TGCAGACACAAGGGAGCGCTGAATCT..................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................CGAGAATTGACAGACtgag.............................................................. | 19 | tgag | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .....................................TACTCTACTGAAGGTAAAGAATGATTCTAGC...................................................................................................................................................................................... | 31 | 2 | 0.50 | 0.50 | - | 0.50 | - | - | - | - | - | - | - | - | - | - |
| .............................................TGAAGGTAAAGAATGATTCTAGCAATTCC................................................................................................................................................................................ | 29 | 2 | 0.50 | 0.50 | - | - | 0.50 | - | - | - | - | - | - | - | - | - |
| ..........................................TACTGAAGGTAAAGAATGATTCTAGCAATT.................................................................................................................................................................................. | 30 | 2 | 0.50 | 0.50 | - | 0.50 | - | - | - | - | - | - | - | - | - | - |
| ..........................................TACTGAAGGTAAAGAATGATTCTAGCAAT................................................................................................................................................................................... | 29 | 2 | 0.50 | 0.50 | - | 0.50 | - | - | - | - | - | - | - | - | - | - |
| ...........................................ACTGAAGGTAAAGAATGATTCTAGCAAT................................................................................................................................................................................... | 28 | 2 | 0.50 | 0.50 | - | - | 0.50 | - | - | - | - | - | - | - | - | - |
| ..........................................TACTGAAGGTAAAGAATGATTCTAGC...................................................................................................................................................................................... | 26 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | 0.50 | - | - | - | - |
| .....................................TACTCTACTGAAGGTAAAGAATGATTCTAG....................................................................................................................................................................................... | 30 | 2 | 0.50 | 0.50 | - | 0.50 | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................TGTCCAAACGAACACCAGACTTTTA.. | 25 | 9 | 0.11 | 0.11 | - | - | - | - | - | - | - | - | - | 0.11 | - | - |
| ..............................................................................................................................................................................................................................TTGTCCAAACGAACACCAGACTTTTA.. | 26 | 9 | 0.11 | 0.11 | - | - | - | - | - | - | - | - | 0.11 | - | - | - |
| ...............................................................................................................................................................................................................................TGTCCAAACGAACACCAGACT...... | 21 | 19 | 0.11 | 0.11 | - | - | - | - | - | - | - | - | - | - | 0.05 | 0.05 |
| ...............................................................................................................................................................................................................................TGTCCAAACGAACACCAGACTTTT... | 24 | 18 | 0.06 | 0.06 | - | - | - | - | - | - | - | - | 0.06 | - | - | - |
| TTATGGTTGAGGTATGCACCCAGCCCATTGATGCCATTACTCTACTGAAGGTAAAGAATGATTCTAGCAATTCCACCCAACACACAGACAAAAGGTGCCATAGACTAACCTCTATTGAGTCTCTGATCTGTAGGGAATGGGTTTCTGATTGCAGGTGAGTAAGGGTCAGCGAGAATTGACAGACAGATGCAGACACAAGGGAGCGCTGAATCTGAATGTATTTTGTCCAAACGAACACCAGACTTTTAAT |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR363960(GSM822762) Adult Library#1Small RNA Miwi IPDuplexed run:. (testes) | SRR363959(GSM822761) AdultSmall RNA Miwi IPread_length: 36. (testes) | mjTestesWT4() Testes Data. (testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................TCTACTGAAGGTAAAGAATGATTCTAGCA..................................................................................................................................................................................... | 29 | 2 | 0.50 | 0.50 | - | - | 0.50 | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................ATTTTGTCCAAACGAACACCAGACTTT.... | 27 | 7 | 0.14 | 0.14 | - | - | 0.14 | - | - | - | - | - | - | - | - | - |