
| Gene: Fkbp8 | ID: uc009map.1_intron_4_0_chr8_73055642_f.5p | SPECIES: mm9 |
 |
 |
|
(5) OTHER.mut |
(6) PIWI.ip |
(1) PIWI.mut |
(1) TDRD1.ip |
(24) TESTES |
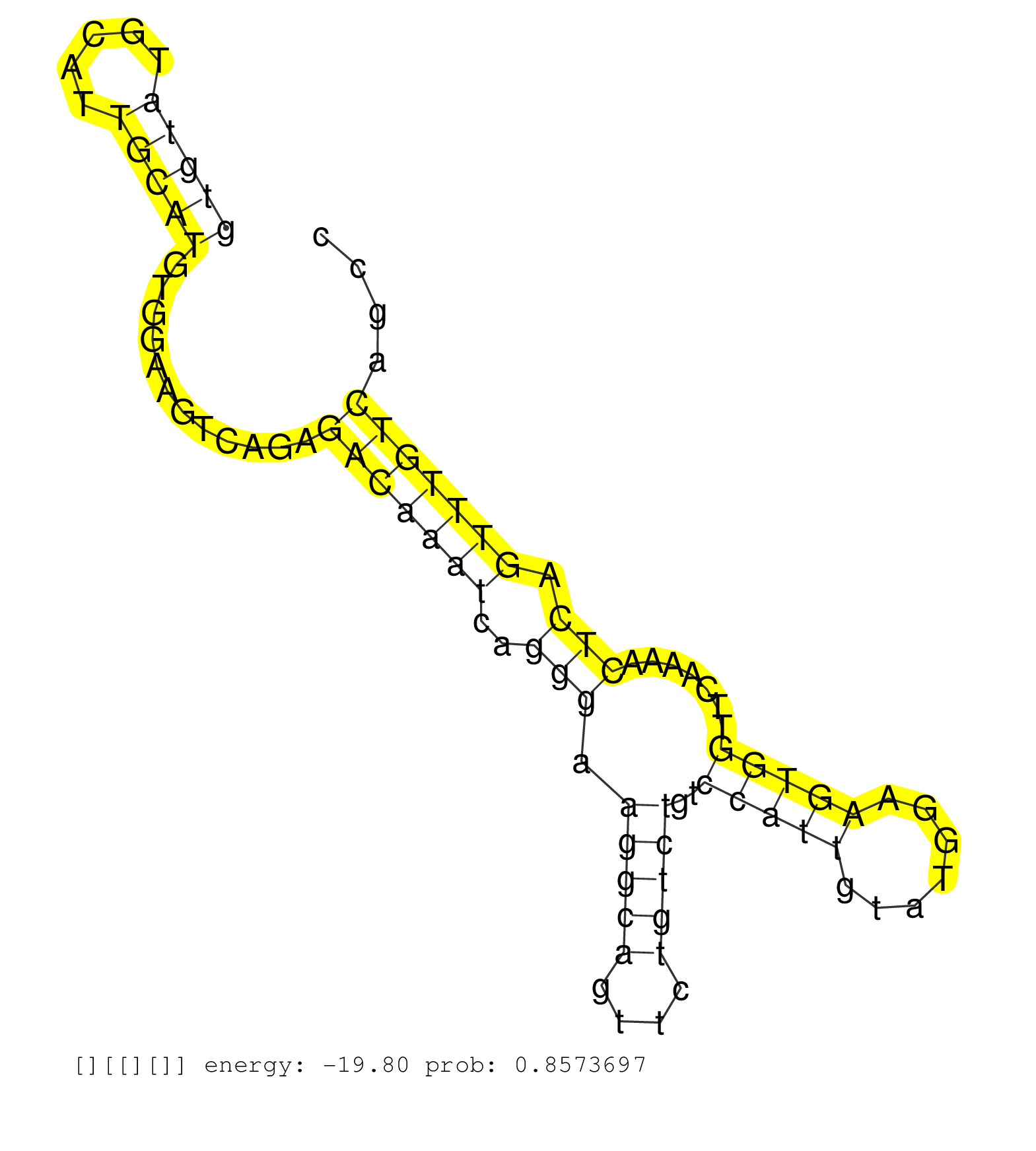
| CGCCAATTCCTATGACCTGGCCATCAAGGCTATCACCTCCAACACCAAAGGTGACCAGTTACCTGGGGTGTGTGTATGCATTGCATGTGGAAGTCAGAGACAAATCAGGGAAGGCAGTTCTGTCTGTCCATTGTATGGAAGTGGTTCAAAACTCAGTTTGTCAGCCTTTGCAGCAAGGACGTTTTCCTGCTGAGCCATCTTGCCAGCCTGTAGGATGGCCAGACTTCTGCACCATCATTTGGGCCATAGA .......................................................................(((((.....)))))............(((((((..(((.(((((....)))))..(((((.......))))).......))).)))))))........................................................................................ .......................................................................72............................................................................................166.................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM475281(GSM475281) total RNA. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................TGCATTGCATGTGGAAGTCAGAGAC..................................................................................................................................................... | 25 | 1 | 5.00 | 5.00 | - | - | 2.00 | - | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................TGGAAGTGGTTCAAAACTCAGTTTGTC........................................................................................ | 27 | 1 | 3.00 | 3.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................TGCATTGCATGTGGAAGTCAGAGACAAA.................................................................................................................................................. | 28 | 1 | 3.00 | 3.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ...................................................................................CATGTGGAAGTCAGAGACAAATCAGGG............................................................................................................................................ | 27 | 1 | 3.00 | 3.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................TGCATTGCATGTGGAAGTCAGAGACA.................................................................................................................................................... | 26 | 1 | 3.00 | 3.00 | - | 1.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................GACAAATCAGGGAAGGCAGTTCTGTC.............................................................................................................................. | 26 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................CATTGCATGTGGAAGTCAGAGACAAA.................................................................................................................................................. | 26 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................GGAAGTCAGAGACAAATCAGGGAAGG........................................................................................................................................ | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................TGGAAGTCAGAGACAAATCAGGGAAGG........................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................GTCAGCCTTTGCAGCAAGGACGTTTTC................................................................ | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................GCATTGCATGTGGAAGTCAGAGACAAATC................................................................................................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................GCATTGCATGTGGAAGTCAGAGAC..................................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................TTGTATGGAAGTGGTTCAAAACTCA............................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .....................................................................................................................................................................................TTTTCCTGCTGAGCCAaagt................................................. | 20 | aagt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..............................................................................CATTGCATGTGGAAGTCAGAGACAAAT................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................ATTGCATGTGGAAGTCAGAG....................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...................................................................................CATGTGGAAGTCAGAGACAAATCAGGGA........................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................TGCATTGCATGTGGAAGTCAGAGACAA................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................TATGCATTGCATGTGGAAGTCAGAGAC..................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................GTTCAAAACTCAGTTTGTCAGCCTTTGC............................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...................................................................................CATGTGGAAGTCAGAGACAAATCAGG............................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................GCATGTGGAAGTCAGAGACAAATCAGG............................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................TGCATTGCATGTGGAAGTCAGAGACAAAT................................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .....................................................................................................................TTCTGTCTGTCCATTGTATGG................................................................................................................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................GCCTTTGCAGCAAGGACGTTTTCCTGt............................................................ | 27 | t | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................TTGTATGGAAGTGGTTCAAAACTCAGT............................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................TCAGCCTTTGCAGCAAGGACGTTTTCCTGCT........................................................... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..................................................................................................................................................................................................................TAGGATGGCCAGACTTCTGta................... | 21 | ta | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................CCTTTGCAGCAAGGACGTTTTCCTGC............................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................GTCAGCCTTTGCAGCAAGGACGTTT.................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................TGGAAGTGGTTCAAAACTCAGTTTGTCAGC..................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .........................................................................................................................................GAAGTGGTTCAAAACTCAGTTTGTCAGC..................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................TGGGGTGTGTGTATGattg........................................................................................................................................................................ | 19 | attg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ........................................................................TGTATGCATTGCATGTGGAAGTCAGAGAC..................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................TATGGAAGTGGTTCAAAACTCAGTTT........................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................ACTCAGTTTGTCAGCCTTTGCAGCAAGGAC...................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................CCTGTAGGATGGCCAGACTTCTGCACC................. | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................GTCAGCCTTTGCAGCAAGGACGTTTT................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................AACTCAGTTTGTCAGCCTTTGC............................................................................... | 22 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................TTGCAGCAAGGACGTTTTCCTG............................................................. | 22 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................TGGAAGTGGTTCAAAACTCAGTTTGTCA....................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................TGGAAGTCAGAGACAAATCAGGG............................................................................................................................................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................GAAGTGGTTCAAAACTCAGTTTGTCAG...................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................TTGTATGGAAGTGGTTCAAAACTCAGTT............................................................................................ | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................TACCTGGGGTGTGTGTATG............................................................................................................................................................................ | 19 | 2 | 0.50 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CGCCAATTCCTATGACCTGGCCATCAAGGCTATCACCTCCAACACCAAAGGTGACCAGTTACCTGGGGTGTGTGTATGCATTGCATGTGGAAGTCAGAGACAAATCAGGGAAGGCAGTTCTGTCTGTCCATTGTATGGAAGTGGTTCAAAACTCAGTTTGTCAGCCTTTGCAGCAAGGACGTTTTCCTGCTGAGCCATCTTGCCAGCCTGTAGGATGGCCAGACTTCTGCACCATCATTTGGGCCATAGA .......................................................................(((((.....)))))............(((((((..(((.(((((....)))))..(((((.......))))).......))).)))))))........................................................................................ .......................................................................72............................................................................................166.................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | GSM475281(GSM475281) total RNA. (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR363957(GSM822759) P20-WTSmall RNA Miwi IPread_length: 36. (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................................................................................TTCCTGCTGAGCCAtagt..................................................... | 18 | tagt | 2.00 | 0.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................AAGGCAGTTCTGTCTGTCCATTGTATGG................................................................................................................ | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |