
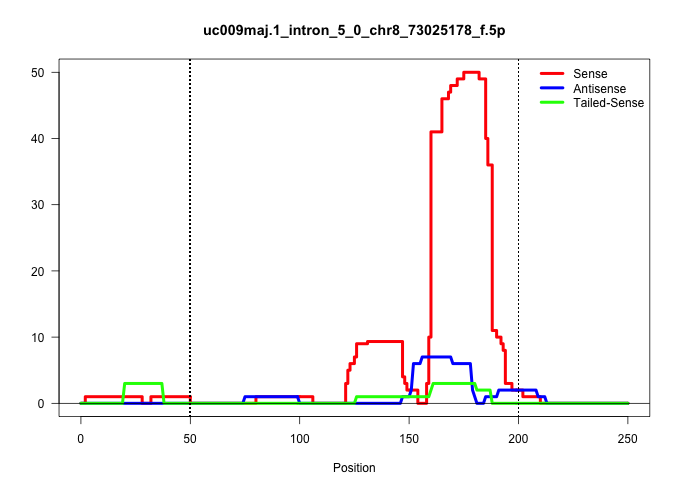
| Gene: Crlf1 | ID: uc009maj.1_intron_5_0_chr8_73025178_f.5p | SPECIES: mm9 |
 |
 |
|
(4) OTHER.mut |
(1) PIWI.mut |
(15) TESTES |
| AATCTGGAGCGAGTGGAGCCACCCCACCGCTGCCTCCACCCCTCGAAGTGGTGAGCACCTCTCCAGGGCTGGCTGGCCCATGGAATCCCCAATCCATCCTGTTCCTCCCCCCCCCCACCCTTTTTTTGAGACAGCGTCTTCAGGTAGCGCATGCTGGCCTTAAATTCAGTATGTAGTCAAGGATGACCTCGAGCTCCTGGTCTTTTTGTCTCCACTTAGAGACAATGGCCAGTGGCCATCACCACCTTTG .............................................................................................................................(((((....((((((......(((((((((((........))))))))).))..))))))..))))).......................................................... ....................................................................................................................117........................................................................192........................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................................................................................TAAATTCAGTATGTAGTCAAGGATGACC.............................................................. | 28 | 1 | 19.00 | 19.00 | 12.00 | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................ACCCCACCGCTGCCacca.................................................................................................................................................................................................................... | 18 | acca | 6.00 | 0.00 | - | - | 3.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - |
| ................................................................................................................................................................TAAATTCAGTATGTAGTCAAGGATG................................................................. | 25 | 1 | 6.00 | 6.00 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................TAAATTCAGTATGTAGTCAAGGATGA................................................................ | 26 | 1 | 5.00 | 5.00 | 3.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...............................................................................................................................................................TTAAATTCAGTATGTAGTCAAGGATG................................................................. | 26 | 1 | 3.00 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................TCAGTATGTAGTCAAGGATGACCTCGAGC........................................................ | 29 | 1 | 3.00 | 3.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................TTAAATTCAGTATGTAGTCAAGGATGACC.............................................................. | 29 | 1 | 3.00 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................TTTTTTGAGACAGCGTCTTCAGGTAG....................................................................................................... | 26 | 1 | 3.00 | 3.00 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................CTTAAATTCAGTATGTAGTCAAGGATGACC.............................................................. | 30 | 1 | 3.00 | 3.00 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................TGAGACAGCGTCTTCAGGTAGCGCATGC................................................................................................ | 28 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................TAAATTCAGTATGTAGTCAAGG.................................................................... | 22 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................GTATGTAGTCAAGGATGACCTCGAGC........................................................ | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..TCTGGAGCGAGTGGAGCCACCCCACC.............................................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................TGACCTCGAGCTCCTGGTCTTTTTGTC........................................ | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................TTTTTGAGACAGCGTCTTCAGGTAG....................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................TGGAATCCCCAATCCATCCTGTTCCT................................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................TTTTGAGACAGCGTCTTCAGGTAG....................................................................................................... | 24 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................AAATTCAGTATGTAGTCAAGGATGtcc.............................................................. | 27 | tcc | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................TGAGACAGCGTCTTCAGGTAGCGCATGt................................................................................................ | 28 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .........................................................................................................................................................................TATGTAGTCAAGGATGACCTCGAGCTCC..................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................TCAGTATGTAGTCAAGGATGACCTCGA.......................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................CCTCCACCCCTCGAAGTG........................................................................................................................................................................................................ | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..........................................................................................................................TTTTTGAGACAGCGTCTTCAGGTAGC...................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................TAAATTCAGTATGTAGTCAAGGATGACCTC............................................................ | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................TCAGTATGTAGTCAAGGATGACCTCGAG......................................................... | 28 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................TTAAATTCAGTATGTAGTCAAGGA................................................................... | 24 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................TTGAGACAGCGTCTTCAGGTAGCG..................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................TGGCCTTAAATTCAGTATGTAGTtaag..................................................................... | 27 | taag | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................GTAGTCAAGGATGACCTCGAGC........................................................ | 22 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................TAAATTCAGTATGTAGTCAAGGATGACa.............................................................. | 28 | a | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................GTCAAGGATGACCTCGAGCTCCTGGTC................................................ | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................CAGCGTCTTCAGGTAG....................................................................................................... | 16 | 3 | 0.33 | 0.33 | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................AGTATGTAGTCAAGGAT.................................................................. | 17 | 5 | 0.20 | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 |
| AATCTGGAGCGAGTGGAGCCACCCCACCGCTGCCTCCACCCCTCGAAGTGGTGAGCACCTCTCCAGGGCTGGCTGGCCCATGGAATCCCCAATCCATCCTGTTCCTCCCCCCCCCCACCCTTTTTTTGAGACAGCGTCTTCAGGTAGCGCATGCTGGCCTTAAATTCAGTATGTAGTCAAGGATGACCTCGAGCTCCTGGTCTTTTTGTCTCCACTTAGAGACAATGGCCAGTGGCCATCACCACCTTTG .............................................................................................................................(((((....((((((......(((((((((((........))))))))).))..))))))..))))).......................................................... ....................................................................................................................117........................................................................192........................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM475281(GSM475281) total RNA. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | SRR037902(GSM510438) testes_rep3. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................................................GCTGGCCTTAAATTCAGTATGTAGTCA....................................................................... | 27 | 1 | 5.00 | 5.00 | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...................................................................................................................................................CGCATGCTGGCCTTAAATTCAGT................................................................................ | 23 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................ACCTCGAGCTCCTGGTCTTTTTGTCTCC..................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................AGCTCCTGGTCTTTTTGT......................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...........................................................................GCCCATGGAATCCCCAATCCATCCT...................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................TTGAGACAGCGTccgt................................................................................................................. | 16 | ccgt | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................TGCTGGCCTTAAATgcgt..................................................................................... | 18 | gcgt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ............................................................................................................................................................GCCTTAAATTCAGTATGTAGTCAAG..................................................................... | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................TGCTGGCCTTAAATTCAGTATGTAGTCAA...................................................................... | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |