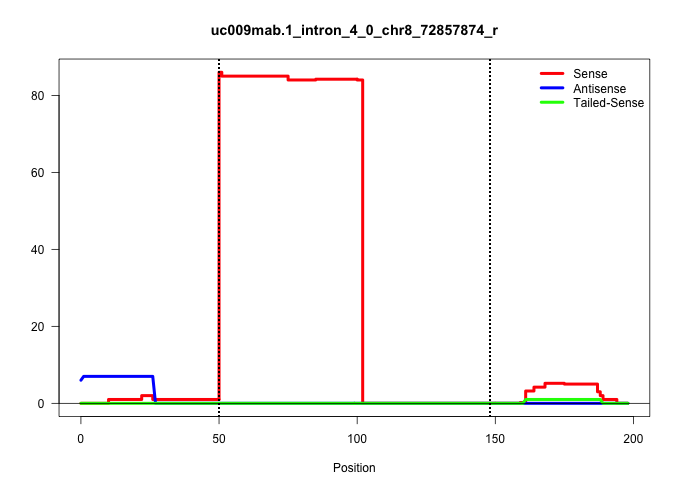
| Gene: Upf1 | ID: uc009mab.1_intron_4_0_chr8_72857874_r | SPECIES: mm9 |
 |
|
(1) PIWI.ip |
(1) PIWI.mut |
(11) TESTES |
| GCCTCATGCAGTTCAGCAAGCCTCGCAAACTTGTCAACACTGTCAACCCGGTGGGCCTAGGGGCTGGCCTGGGACCTGGGAAAGCTGGGAGCTCTTAGCCTCTGCTGCTGTTGACTGACACATTCCTCACTCTTATGCCACCCTGCAGGGTGCCCGCTTCATGACTACTGCCATGTACGATGCCCGTGAGGCCATCAT |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | mjTestesWT3() Testes Data. (testes) | mjTestesWT2() Testes Data. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTGGGCCTAGGGGCTGGCCTGGGACCTGGGAAAGCTGGGAGCTCTTAGCCTC................................................................................................ | 52 | 1 | 84.00 | 84.00 | 84.00 | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................TGACTACTGCCATGTACGATGCCCGT........... | 26 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................TGACTACTGCCATGTACGATGCCCGTGt......... | 28 | t | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..........GTTCAGCAAGCCTCGC............................................................................................................................................................................ | 16 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..................................................GTGGGCCTAGGGGCTGGCCTGGGAC........................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ......................TCGCAAACTTGTCAACACTGTCAACCCGG................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .................................................................................................................................................................TGACTACTGCCATGTACGATGCCCGTG.......... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................CTACTGCCATGTACGATGCCCGTGA......... | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................TGCCATGTACGATGCCCGTGAGGCCA.... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .....................................................................................TGGGAGCTCTTAGCC.................................................................................................. | 15 | 5 | 0.20 | 0.20 | - | - | - | - | - | - | - | - | - | - | 0.20 |
| ...............................................................................................................................................................CATGACTACTGCCATG....................... | 16 | 5 | 0.20 | 0.20 | - | - | - | - | 0.20 | - | - | - | - | - | - |
| GCCTCATGCAGTTCAGCAAGCCTCGCAAACTTGTCAACACTGTCAACCCGGTGGGCCTAGGGGCTGGCCTGGGACCTGGGAAAGCTGGGAGCTCTTAGCCTCTGCTGCTGTTGACTGACACATTCCTCACTCTTATGCCACCCTGCAGGGTGCCCGCTTCATGACTACTGCCATGTACGATGCCCGTGAGGCCATCAT |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363964(GSM822766) AdultGlobal 5'-RACEread_length: 105. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | mjTestesWT3() Testes Data. (testes) | mjTestesWT2() Testes Data. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .CCTCATGCAGTTCAGCAAGCCTCGCA........................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |