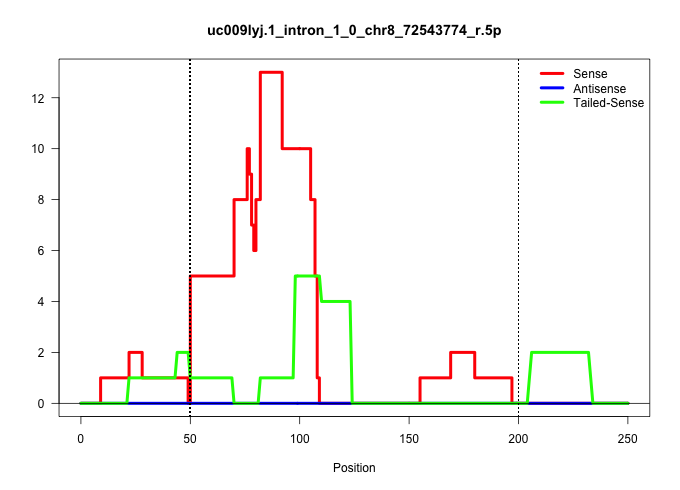
| Gene: mKIAA0892 | ID: uc009lyj.1_intron_1_0_chr8_72543774_r.5p | SPECIES: mm9 |
 |
|
(1) OTHER.mut |
(5) PIWI.ip |
(13) TESTES |
| CAAGATCCCAGACATGTCTGTGCAGCTGTGGTCATCTGCCCTCCTGAGAGGTGAGAGTGAACAAAGCTGCCAGGCATGCTTGTAGGGCCCTGACATCATACAAGCAGCGGTGTATCTCCAGGGTTTTTTTTTATTTTTGTTTGTTTATTTATTGGAAACAGGGTCTAGCTGGCCCGGAACTCTGGGTAGACAAGGCTGGTCTTTTTTTTTTGGTTTTTCGAGACAGGGTTTCCCTGAATAGCTTTGGCTG |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesWT1() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................TACAAGCAGCGGTGTATCTCCAGagt.............................................................................................................................. | 26 | agt | 4.00 | 0.00 | - | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................TAGGGCCCTGACATCATACAAGCAGC.............................................................................................................................................. | 26 | 1 | 4.00 | 4.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................CAGGCATGCTTGTAGGGCCCTG.............................................................................................................................................................. | 22 | 1 | 3.00 | 3.00 | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGAGTGAACAAAGCTGCCAGGCATG............................................................................................................................................................................ | 28 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ................................................................................TGTAGGGCCCTGACATCATACAAGCAG............................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..................................................GTGAGAGTGAACAAAGCTGCCAGGCATGC........................................................................................................................................................................... | 29 | 1 | 2.00 | 2.00 | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - |
| ............................................................................TGCTTGTAGGGCCCTGACATCATACAAGC................................................................................................................................................. | 29 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................TTGTAGGGCCCTGACATCATACAAGCAG............................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGAGTGAACAAAGCTGCCAGGCAT............................................................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ............................................TGAGAGGTGAGAGTGAACAAAGCTGt.................................................................................................................................................................................... | 26 | t | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................TAGGGCCCTGACATCATACAAGCAGCGt............................................................................................................................................ | 28 | t | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .........AGACATGTCTGTGCAGCTG.............................................................................................................................................................................................................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..................................................................................TAGGGCCCTGACATCATACAAGCAGCG............................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................AAACAGGGTCTAGCTGGCCCGGAAC...................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .............................................................................................................................................................................................................TTTTTTGGTTTTTCGAGACAGGGTTTaa................. | 28 | aa | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ......................CAGCTGTGGTCATCTGCCCTCCTGAGAa........................................................................................................................................................................................................ | 28 | a | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ......................CAGCTGTGGTCATCTGCCCTCCTGAGA......................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .........................................................................................................................................................................TGGCCCGGAACTCTGGGTAGACAAGGCT..................................................... | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................TTTTTGGTTTTTCGAGACAGGGTTTata................ | 28 | ata | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| CAAGATCCCAGACATGTCTGTGCAGCTGTGGTCATCTGCCCTCCTGAGAGGTGAGAGTGAACAAAGCTGCCAGGCATGCTTGTAGGGCCCTGACATCATACAAGCAGCGGTGTATCTCCAGGGTTTTTTTTTATTTTTGTTTGTTTATTTATTGGAAACAGGGTCTAGCTGGCCCGGAACTCTGGGTAGACAAGGCTGGTCTTTTTTTTTTGGTTTTTCGAGACAGGGTTTCCCTGAATAGCTTTGGCTG |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesWT1() Testes Data. (testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | GSM509276(GSM509276) small RNA cloning by length. (testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR037902(GSM510438) testes_rep3. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................CTGACATCATACAAaagg................................................................................................................................................... | 18 | aagg | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |