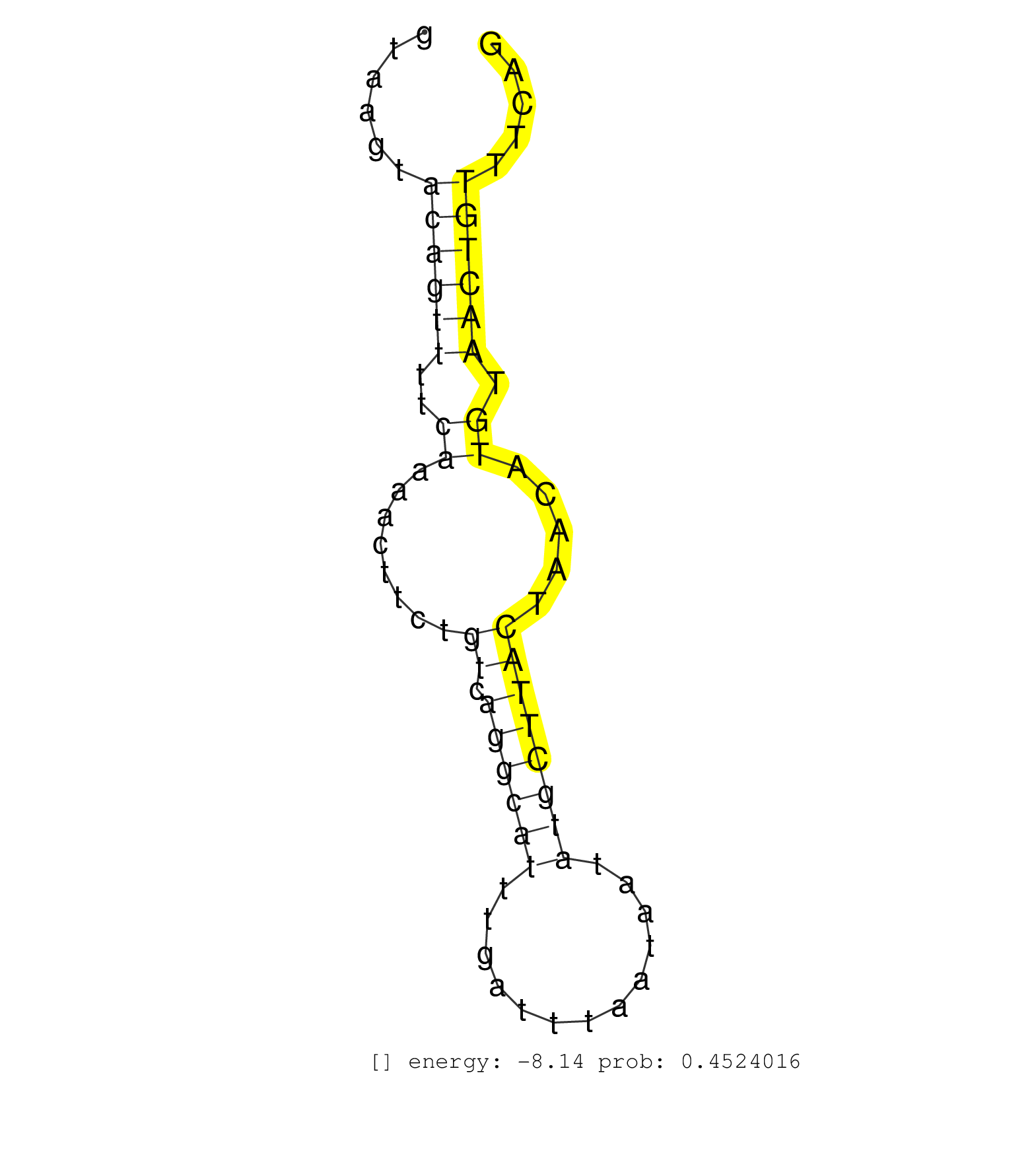

| Gene: Nek1 | ID: uc009ltt.1_intron_21_0_chr8_63568295_f | SPECIES: mm9 |
 |
 |
|
(6) OTHER.mut |
(3) PIWI.ip |
(17) TESTES |
| TAACTGATACCCAGGAAGAAGAAATGGAAAAGAGTAACAGTGCTATTTCAGTAAGTACAGTTTTCAAAACTTCTGTCAGGCATTTGATTTAATAATATGCTTACTAACATGTAACTGTTTCAGAGTAAGCGAGAAATCCTGCGTAGGCTAAATGAAAATCTTAAAGCTCAAGA ........................................................((((((..((........((.((((((.............)))))))).....)).))))))....................................................... ..................................................51......................................................................123................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | GSM475281(GSM475281) total RNA. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................CTTACTAACATGTAACTGTTTCAGA................................................. | 25 | 1 | 20.00 | 20.00 | 7.00 | 6.00 | 2.00 | - | 1.00 | 2.00 | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 |
| ..................................................................................................GCTTACTAACATGTAACTGTTTCAGA................................................. | 26 | 1 | 7.00 | 7.00 | 2.00 | - | 1.00 | - | 2.00 | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - |
| ..................................................................................................GCTTACTAACATGTAACTGTTTCAG.................................................. | 25 | 1 | 5.00 | 5.00 | - | - | 2.00 | 1.00 | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - |
| ...................................................................................................CTTACTAACATGTAACTGTTTCAGAa................................................ | 26 | a | 3.00 | 20.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................CTTACTAACATGTAACTGTTTCAG.................................................. | 24 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................ATGCTTACTAACATGTAACTGTTTCAG.................................................. | 27 | 1 | 2.00 | 2.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................GCTTACTAACATGTAACTGTTTCAGAa................................................ | 27 | a | 2.00 | 7.00 | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ................................................................................................ATGCTTACTAACATGTAACTGTTTCAGA................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..................................................................................................GCTTACTAACATGTAACTGTTTCAGAaa............................................... | 28 | aa | 1.00 | 7.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..................................................................................................GCTTACTAACATGTAACTGTTTCAGg................................................. | 26 | g | 1.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ........................TGGAAAAGAGTAACAGTGCTATTTCAagt........................................................................................................................ | 29 | agt | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .................GAAGAAATGGAAAAGAGTAACAGTGC.................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..................................................................................................GCTTACTAACATGTAACTGTTTCAGAG................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................CTTACTAACATGTAACTGTTTCAGt................................................. | 25 | t | 1.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................TAGGCTAAATGAAAATCTTAAAGCTCAAGA | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..................................................................................................GCTTACTAACATGTAACTGTTTCAt.................................................. | 25 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| TAACTGATACCCAGGAAGAAGAAATGGAAAAGAGTAACAGTGCTATTTCAGTAAGTACAGTTTTCAAAACTTCTGTCAGGCATTTGATTTAATAATATGCTTACTAACATGTAACTGTTTCAGAGTAAGCGAGAAATCCTGCGTAGGCTAAATGAAAATCTTAAAGCTCAAGA ........................................................((((((..((........((.((((((.............)))))))).....)).))))))....................................................... ..................................................51......................................................................123................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | GSM475281(GSM475281) total RNA. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM475279(GSM475279) Miwi-IP. (miwi testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|