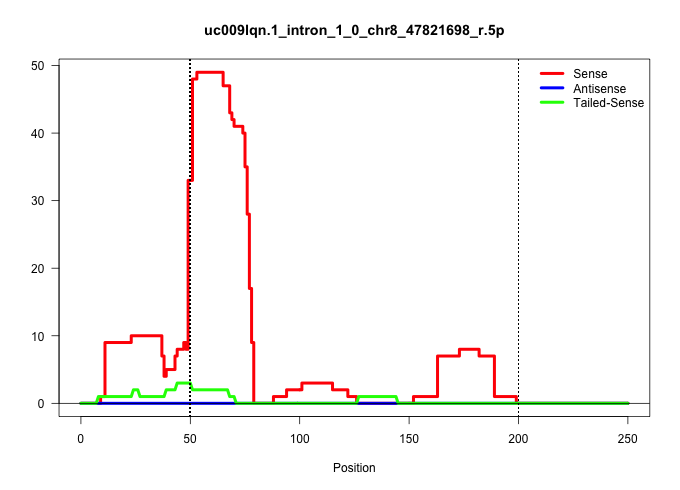
| Gene: AK144632 | ID: uc009lqn.1_intron_1_0_chr8_47821698_r.5p | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(5) PIWI.ip |
(2) PIWI.mut |
(24) TESTES |
| CAGGCTGCATATACAAACTCATCTTGGAAAGTCGCCCGCTGCAAAGGAATGTAAGGAACGAAGTCAGCTGAAAAGACGGCATTACTTGTGCCCCTACTTTATTGGATCTCCTACTGGGGAACGGGGGGGGGGGGGGGGTTTGAGGGGGGTTGAAGGGCCAGCCTGGCACTCAGTGTGGAAGCTAGGATGGTTAGGAAGGCCTTCCCCTAGAAAGTCACTCTCCAAAGTTACTTCATGTCTTTGCCATAAC ..........................................................(((..((.((((......))))....((((.((((((.(((..(((..(((((....))))).)))..)))....)))))).)))).))..)))...(((((.((((((..((.....))..)))))).))))).......................................................... ............................................45..................................................................................................................................................193....................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesWT4() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | mjTestesWT1() Testes Data. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | mjTestesWT3() Testes Data. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR037903(GSM510439) testes_rep4. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................TGTAAGGAACGAAGTCAGCTGAAAAG............................................................................................................................................................................... | 26 | 1 | 10.00 | 10.00 | - | 2.00 | - | - | 1.00 | - | 5.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................TGTAAGGAACGAAGTCAGCTGAAAAGACGG........................................................................................................................................................................... | 30 | 1 | 9.00 | 9.00 | - | - | 2.00 | - | - | - | - | - | 1.00 | 4.00 | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .................................................TGTAAGGAACGAAGTCAGCTGAAAAGAC............................................................................................................................................................................. | 28 | 1 | 8.00 | 8.00 | 3.00 | 1.00 | - | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ...................................................TAAGGAACGAAGTCAGCTGAAAAGACG............................................................................................................................................................................ | 27 | 1 | 7.00 | 7.00 | 1.00 | 2.00 | - | - | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .................................................TGTAAGGAACGAAGTCAGCTGAAAAGA.............................................................................................................................................................................. | 27 | 1 | 7.00 | 7.00 | 2.00 | - | - | 2.00 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................TGGCACTCAGTGTGGAAGCTAGGATG............................................................. | 26 | 1 | 6.00 | 6.00 | 4.00 | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TAAGGAACGAAGTCAGCTGAAAAGACGG........................................................................................................................................................................... | 28 | 1 | 6.00 | 6.00 | 1.00 | 1.00 | - | - | - | - | - | 2.00 | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...........TACAAACTCATCTTGGAAAGTCGCCCG.................................................................................................................................................................................................................... | 27 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - |
| .......................................TGCAAAGGAATGTAAGGAACGAAGTCAGC...................................................................................................................................................................................... | 29 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TAAGGAACGAAGTCAGCTGAAAAGAC............................................................................................................................................................................. | 26 | 1 | 3.00 | 3.00 | - | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........TACAAACTCATCTTGGAAAGTCGCCCGC................................................................................................................................................................................................................... | 28 | 1 | 3.00 | 3.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - |
| ...........TACAAACTCATCTTGGAAAGTCGCCC..................................................................................................................................................................................................................... | 26 | 1 | 2.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................AAGGGCCAGCCTGGCACTCAGTGTGGAAGC.................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................TGTAAGGAACGAAGTCAGCTGAAAAGACG............................................................................................................................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................TGGAAAGTCGCCCGCTGCAAAGGcatg....................................................................................................................................................................................................... | 27 | catg | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................TGCAAAGGAATGTAAGGAACGAAGTC......................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................TGTAAGGAACGAAGTCAGCTGAAAA................................................................................................................................................................................ | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........TATACAAACTCATCTTGGAAAGTCGCCC..................................................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................TGCAAAGGAATGTAAGGAACGAAGTCAGt...................................................................................................................................................................................... | 29 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................AGGAACGAAGTCAGCTGAAAAGACGG........................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................GGGGGGGGGGGTTTttga......................................................................................................... | 18 | ttga | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...........................................AAGGAATGTAAGGAACGAAGTCAGC...................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................AGGAATGTAAGGAACGAAGTCAGCTGt................................................................................................................................................................................... | 27 | t | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................TGTGGAAGCTAGGATGGTTAGGAAGG................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................TACTTTATTGGATCTCCTACTGGGGAAC................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ........ATATACAAACTCATCggtc............................................................................................................................................................................................................................... | 19 | ggtc | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...............................................AATGTAAGGAACGAAGTC......................................................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................TGCAAAGGAATGTAAGGAACG.............................................................................................................................................................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ........................................................................................TGCCCCTACTTTATTGGATCTCCTACT....................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...........................................AAGGAATGTAAGGAACGAAGTCAGCT..................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................TTGGATCTCCTACTGGGGAACGGGG............................................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................TTGGAAAGTCGCCCGCTGCAAAGGA.......................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ............................................AGGAATGTAAGGAACGAAGTCAGCTG.................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CAGGCTGCATATACAAACTCATCTTGGAAAGTCGCCCGCTGCAAAGGAATGTAAGGAACGAAGTCAGCTGAAAAGACGGCATTACTTGTGCCCCTACTTTATTGGATCTCCTACTGGGGAACGGGGGGGGGGGGGGGGTTTGAGGGGGGTTGAAGGGCCAGCCTGGCACTCAGTGTGGAAGCTAGGATGGTTAGGAAGGCCTTCCCCTAGAAAGTCACTCTCCAAAGTTACTTCATGTCTTTGCCATAAC ..........................................................(((..((.((((......))))....((((.((((((.(((..(((..(((((....))))).)))..)))....)))))).)))).))..)))...(((((.((((((..((.....))..)))))).))))).......................................................... ............................................45..................................................................................................................................................193....................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesWT4() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | mjTestesWT1() Testes Data. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR014233(GSM319957) 16.5 dpc MIWI2. (miwi2 testes) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | mjTestesWT3() Testes Data. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR037903(GSM510439) testes_rep4. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...CATATACAAACTCAgctt..................................................................................................................................................................................................................................... | 18 | gctt | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |