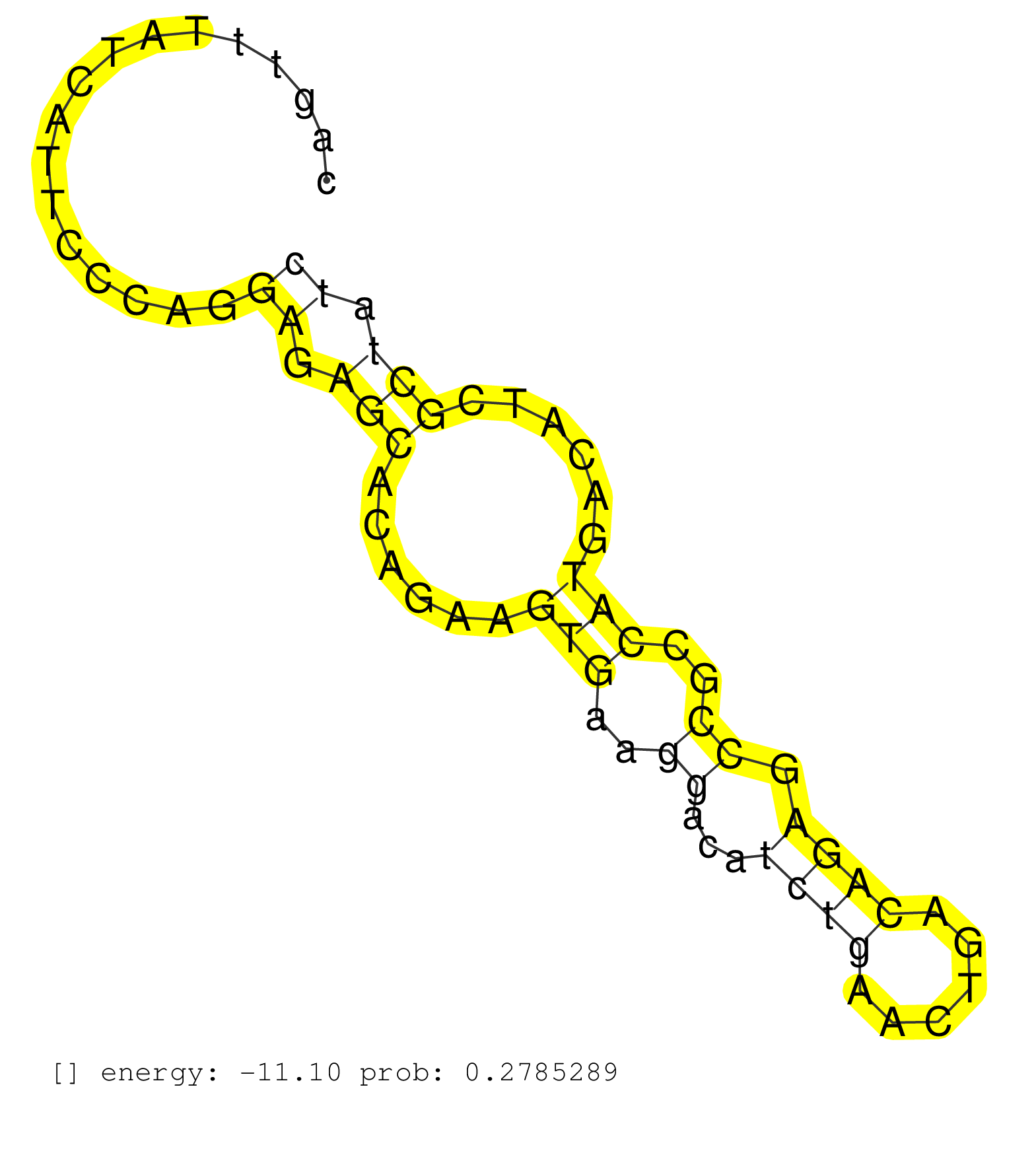

| Gene: Prosc | ID: uc009lhu.1_intron_1_0_chr8_28155608_f.3p | SPECIES: mm9 |
 |
 |
|
(4) OTHER.mut |
(5) PIWI.ip |
(1) PIWI.mut |
(15) TESTES |
| AATGACAGTGAATTGATGGGAATGAGAAGGGAACCAGCGCCAGGCTTAGATGCCTCTGCGTTTCCTTGTGTCTCAGTGGGAGATTGGCCGAAGAGAGGGGTGGCAGTTTATCATTCCCAGGAGAGCACAGAAGTGAAGGACATCTGAACTGACAGAGCCGCCATGACATCGCTATCAAACGGTCCCCTTCTTCTCTTCAGATTCTGTCTTCCTGTCCCGAGATCAAGTGGCACTTCATTGGTCATTTACA ........................................................................................................................((.(((......(((..((...((((......)))).))..)))......))).)).......................................................................... .......................................................................................................104.....................................................................176........................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................AACTGACAGAGCCGCCATGACATCGC.............................................................................. | 26 | 1 | 7.00 | 7.00 | 4.00 | - | - | - | 1.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | - |
| .......................................................................................................................................AAGGACATCTGAACTGACAGAGCCGCC........................................................................................ | 27 | 1 | 5.00 | 5.00 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................AACTGACAGAGCCGCCATGACATCGCT............................................................................. | 27 | 1 | 4.00 | 4.00 | - | 1.00 | - | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................TCTGAACTGACAGAGCCGCCATGACATC................................................................................ | 28 | 1 | 3.00 | 3.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................AAGGACATCTGAACTGACAGAGCCGCCA....................................................................................... | 28 | 1 | 3.00 | 3.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..................................................................................................................................................AACTGACAGAGCCGCCATGACATCGCTA............................................................................ | 28 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................ATCTGAACTGACAGAGCCGCCATG..................................................................................... | 24 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................AACTGACAGAGCCGCCATGACATCGCTATC.......................................................................... | 30 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................AAGGACATCTGAACTGACAGAGCCGC......................................................................................... | 26 | 1 | 2.00 | 2.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................GAAGGACATCTGAACTGACAGAGCCGCC........................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................GGACATCTGAACTGACAGAGCCGCCAT...................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................AACTGACAGAGCCGCCATGACATCGCTt............................................................................ | 28 | t | 1.00 | 4.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................CTGAACTGACAGAGCCGCCATGACATCGC.............................................................................. | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......GTGAATTGATGGGAATGAG................................................................................................................................................................................................................................ | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..............................................................................................................................................................................................TTCTCTTCAGATTCTGTCTTCCTGTC.................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ....................................................................................................................................................................................................................TGTCCCGAGATCAAGTGGCACTTCATTGGTt....... | 31 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...........................................................................................................................AGCACAGAAGTGAAGGACATCTGAACTG................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ........................................................................................................................................AGGACATCTGAACTGACAGAGCCGCC........................................................................................ | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................GACATCTGAACTGACAGAGCCGCCATGACATC................................................................................ | 32 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................TCCCGAGATCAAGTGGCACTTCAT............ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..................................................................................................................................................AACTGACAGAGCCGCCATGACATCGCTAT........................................................................... | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................GACATCTGAACTGACAGAGCCGCCATGACt.................................................................................. | 30 | t | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................TATCATTCCCAGGAGAGCACAGAAGTG................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| AATGACAGTGAATTGATGGGAATGAGAAGGGAACCAGCGCCAGGCTTAGATGCCTCTGCGTTTCCTTGTGTCTCAGTGGGAGATTGGCCGAAGAGAGGGGTGGCAGTTTATCATTCCCAGGAGAGCACAGAAGTGAAGGACATCTGAACTGACAGAGCCGCCATGACATCGCTATCAAACGGTCCCCTTCTTCTCTTCAGATTCTGTCTTCCTGTCCCGAGATCAAGTGGCACTTCATTGGTCATTTACA ........................................................................................................................((.(((......(((..((...((((......)))).))..)))......))).)).......................................................................... .......................................................................................................104.....................................................................176........................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029039(GSM433291) 25dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) |
|---|