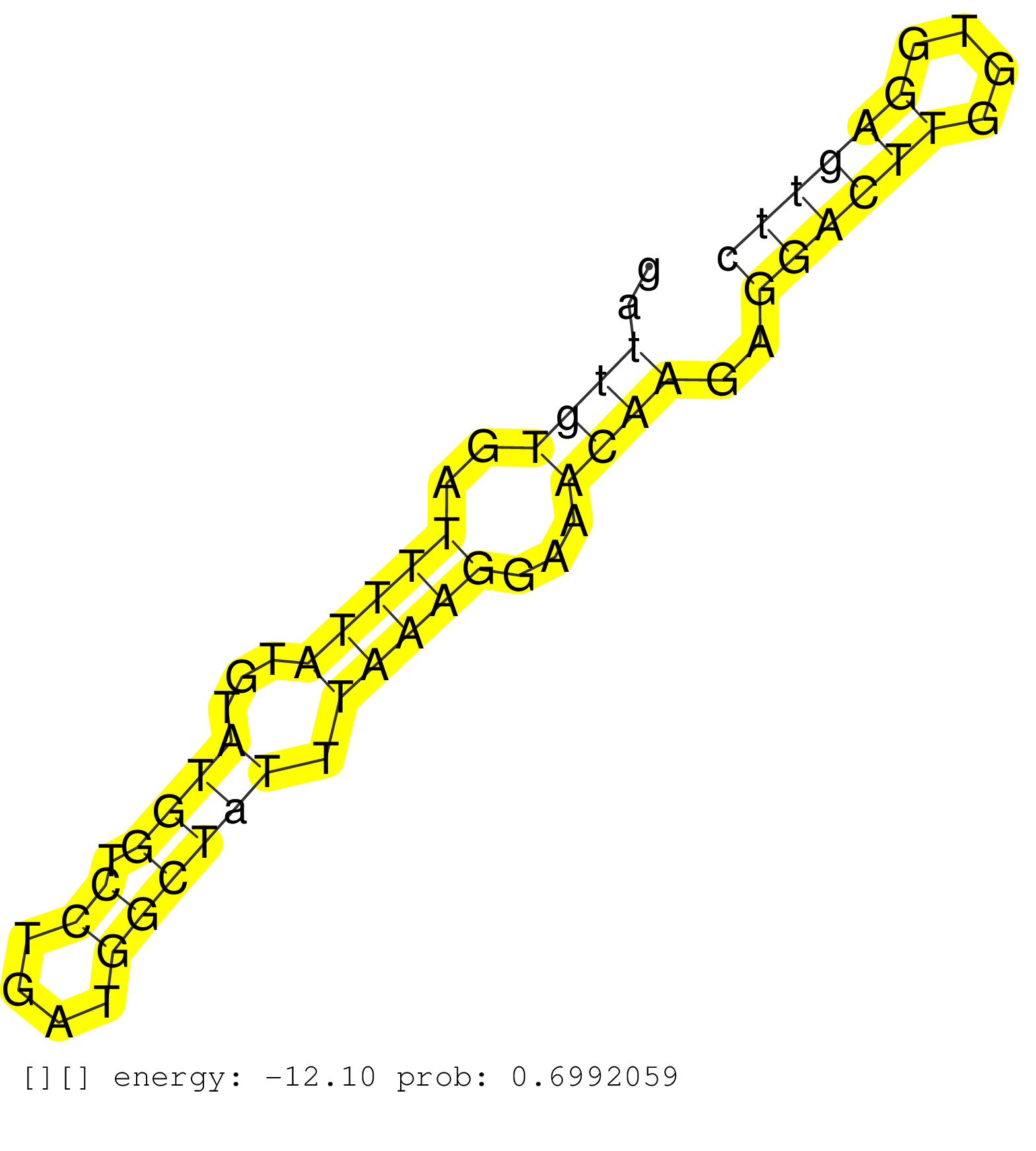
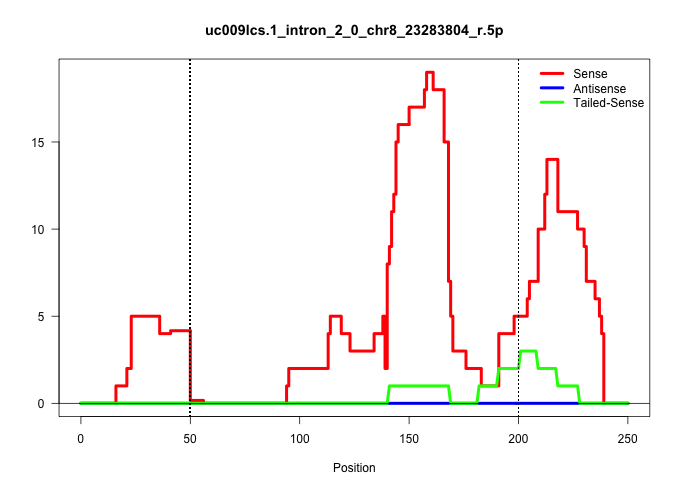
| Gene: Ckap2 | ID: uc009lcs.1_intron_2_0_chr8_23283804_r.5p | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(4) PIWI.ip |
(2) PIWI.mut |
(17) TESTES |
| TTGAAAATATTATCTCAATCTATGAGAAGGCCATCCTGGCAGGGGCTCAGGTAAGATCAAAATGCACCGAGTTGTTTTGGAGCAGCACATCACTGTGCAGTTCGATCCGATTGTGATTTTATGTATGGTCCTGATGGCTATTTAAAGGAAACAAGAGGACTTGGTGGAGTTCAGTGGGAGAGCAGGGGAGATGGTAGCAGCAGAAACAGTGAAGGCTGCAAGGACTGCATTATGCTTTCCAACCTCCTAA ..............................................................................................................((((..(((((...((((.((....)))))).)))))...))))..((((((....)))))).............................................................................. ............................................................................................................109............................................................172............................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................GAGAAGGCCATCCTGGCAGGGGCTCAG........................................................................................................................................................................................................ | 27 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | 2.00 | - | 1.00 | - | - | - | - | - | - |
| ............................................................................................................................................TTTAAAGGAAACAAGAGGACTTGGTGGA.................................................................................. | 28 | 1 | 3.00 | 3.00 | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................TGGTAGCAGCAGAAACAGTGAAGGCTG................................ | 27 | 1 | 3.00 | 3.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................TTTAAAGGAAACAAGAGGACTTGGTG.................................................................................... | 26 | 1 | 3.00 | 3.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................TGATTTTATGTATGGTCCTGATGGCT............................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................AAGGAAACAAGAGGACTTGGTGGAGT................................................................................ | 26 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................AGGCTGCAAGGACTGCATTATGCTTTC........... | 27 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................TAAAGGAAACAAGAGGACTTGGTGGA.................................................................................. | 26 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................GGCTGCAAGGACTGCATTATGCTTTC........... | 26 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................AAGGAAACAAGAGGACTTGGTGGAG................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .................................................................................................................................................................................................................TGAAGGCTGCAAGGACTGCATTATGCTTT............ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ............................................................................................................................................................................................................AACAGTGAAGGCTGCAAGGACTGCATT................... | 27 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................AAAGGAAACAAGAGGACTTGGTGGA.................................................................................. | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................GGGAGATGGTAGCAGCAGAAACAGT........................................ | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................ACAGTGAAGGCTGCAAGGACTGCATT................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................AATCTATGAGAAGGCCATCC...................................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................ACTTGGTGGAGTTCAGTGGGAGAGC................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................ATGAGAAGGCCATCCTGGCAGGGGCTCAG........................................................................................................................................................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .................................................................................................................................................................................................................TGAAGGCTGCAAGGACTGCATTATGC............... | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................TTAAAGGAAACAAGAGGACTTGGTGGtt................................................................................. | 28 | tt | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................AGGAAACAAGAGGACTTGGTGGAG................................................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................AGAAACAGTGAAGGCTGCAAGGACTGt...................... | 27 | t | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................TGAAGGCTGCAAGGACTGCATTATGCTT............. | 28 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................TGGTAGCAGCAGAAACAGTGAAGGCTt................................ | 27 | t | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................GAAGGCTGCAAGGACTGCAT.................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..................................................................................................................GATTTTATGTATGGTCCTGATGGCT............................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...............................................................................................TGCAGTTCGATCCGATTGTGATTTTATG............................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................GTGCAGTTCGATCCGATTGTGATTT................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................AGCAGAAACAGTGAAGGCTGCAAGGACTG....................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................TGGCTATTTAAAGGAAACAAGAGGACT......................................................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................TATTTAAAGGAAACAAGAGGACTTGGTGGA.................................................................................. | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................ACAAGAGGACTTGGTGGAGTTCAGTG.......................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .............................................................................................................................................................GACTTGGTGGAGTTCAGTGGGAGAGCAG................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................TTAAAGGAAACAAGAGGACTTGGTGGA.................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................CAGGGGAGATGGTAGCAGCAGAAACAa......................................... | 27 | a | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................................GGGGCTCAGGTAAGA.................................................................................................................................................................................................. | 15 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | 0.17 | - | - | - | - | - |
| TTGAAAATATTATCTCAATCTATGAGAAGGCCATCCTGGCAGGGGCTCAGGTAAGATCAAAATGCACCGAGTTGTTTTGGAGCAGCACATCACTGTGCAGTTCGATCCGATTGTGATTTTATGTATGGTCCTGATGGCTATTTAAAGGAAACAAGAGGACTTGGTGGAGTTCAGTGGGAGAGCAGGGGAGATGGTAGCAGCAGAAACAGTGAAGGCTGCAAGGACTGCATTATGCTTTCCAACCTCCTAA ..............................................................................................................((((..(((((...((((.((....)))))).)))))...))))..((((((....)))))).............................................................................. ............................................................................................................109............................................................172............................................................................ |
Size | Perfect hit | Total Norm | Perfect Norm | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR014235(GSM319959) 2 dpp total. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................................................GTTCAGTGGGAGAGCgt................................................................... | 17 | gt | 3.00 | 0.00 | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | 1.00 |
| ...........................................................................................................................................................................................................AGTGAAGGCTGCAAGGaaaa........................... | 20 | aaaa | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |