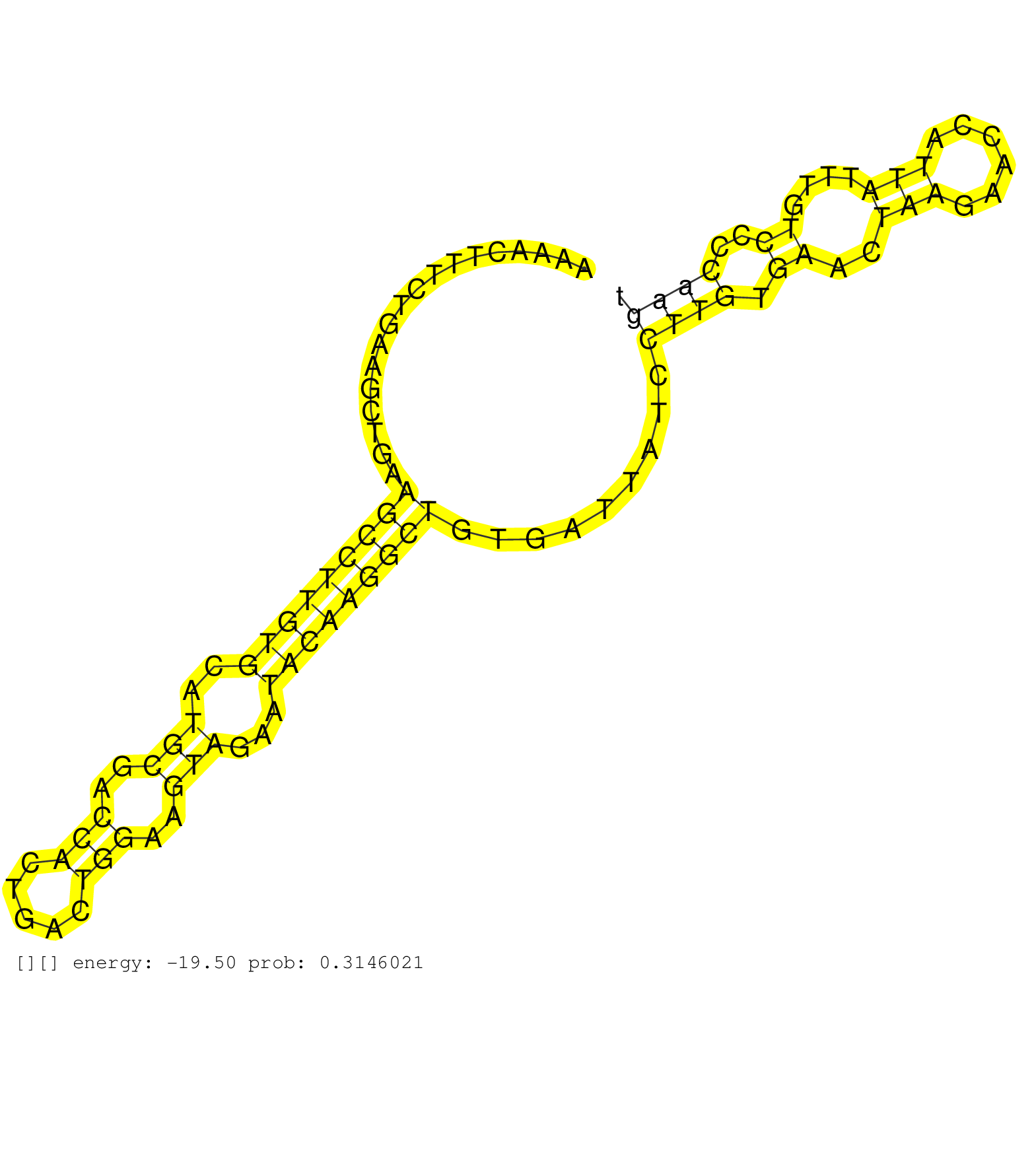
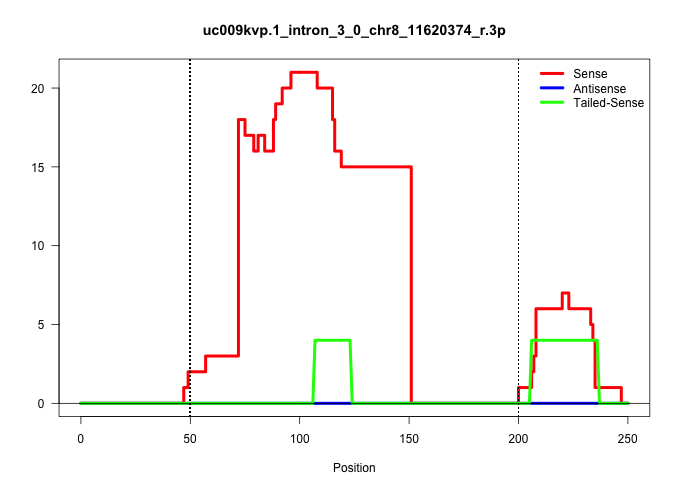
| Gene: Ankrd10 | ID: uc009kvp.1_intron_3_0_chr8_11620374_r.3p | SPECIES: mm9 |
 |
 |
|
(2) OTHER.mut |
(4) PIWI.ip |
(1) PIWI.mut |
(21) TESTES |
| ATATTTCCTTTCTCTCTCTCTTTTTAAGTAGATATGTTTGATGAGTATCTAAAACTTTCTGAAGCTGAAGCCTTGTGCATGCGACCACTGACTGGAAGTAGAATACAAGGCTGTGATTATCCTTGTGAACTAAGAACCATTATTTGTCCCCAAGTCTATAGTTATTTTTAAGTTTGTTTGTTTGTTTGTTGCCTTAACAGTAACCCTAACAAGCGCAAGCATCAGGGTTTGGATTGCTCATCTCTCTGAT ....................................................................(((((((((..(((..(((.....)))..)))...))))))))).........((((.((..(((......)))....))..))))................................................................................................ ..................................................51......................................................................................................155............................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT1() Testes Data. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR363960(GSM822762) Adult Library#1Small RNA Miwi IPDuplexed run:. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR037903(GSM510439) testes_rep4. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................TTGTGCATGCGACCACTGACTGGAAGTAGAATACAAGGCTGTGATTATCCTT.............................................................................................................................. | 52 | 1 | 15.00 | 15.00 | 15.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................GACTGGAAGTAGAATACAAGGCTGTGA...................................................................................................................................... | 27 | 1 | 5.00 | 5.00 | - | 4.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................TAACAAGCGCAAGCATCAGGGTTTGGATTGt............. | 31 | t | 4.00 | 0.00 | - | - | - | - | 3.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................AGGCTGTGATTATtggc.............................................................................................................................. | 17 | tggc | 4.00 | 0.00 | - | - | - | - | - | 3.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................ACAAGCGCAAGCATCAGGGTTTGGATT............... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................TGACTGGAAGTAGAATACAAGGCTG......................................................................................................................................... | 25 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................TGACTGGAAGTAGAATACAAGGCTGTG....................................................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - |
| .........................................................TCTGAAGCTGAAGCCTTGTGCATGCGA...................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ................................................................................................................................................................................................................ACAAGCGCAAGCATCAGGGTTTGGAT................ | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................TGGAAGTAGAATACAAGGCTGTGA...................................................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ................................TATGTTTGATGAGTATCTAAAACTTTCTGAA........................................................................................................................................................................................... | 31 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................CGACCACTGACTGGAAGTAGAATACAA.............................................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ............................................................................................................................................................................................................................ATCAGGGTTTGGATTGCTCATCTCTCT... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ........................................................................................................................................................................................................TAACCCTAACAAGCGCAAGCATC........................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .................................................TAAAACTTTCTGAAGCTGAAGCCTTGTGCA........................................................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................AGTAGAATACAAGGCTGTGATTA................................................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...............................................TCTAAAACTTTCTGAAGCTGAAGCCTTG............................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................AACAAGCGCAAGCATCAGGGTTTGGA................. | 26 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................CGACCACTGACTGGAAGT....................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................TAACAAGCGCAAGCATCAGGGTTTGGATT............... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ATATTTCCTTTCTCTCTCTCTTTTTAAGTAGATATGTTTGATGAGTATCTAAAACTTTCTGAAGCTGAAGCCTTGTGCATGCGACCACTGACTGGAAGTAGAATACAAGGCTGTGATTATCCTTGTGAACTAAGAACCATTATTTGTCCCCAAGTCTATAGTTATTTTTAAGTTTGTTTGTTTGTTTGTTGCCTTAACAGTAACCCTAACAAGCGCAAGCATCAGGGTTTGGATTGCTCATCTCTCTGAT ....................................................................(((((((((..(((..(((.....)))..)))...))))))))).........((((.((..(((......)))....))..))))................................................................................................ ..................................................51......................................................................................................155............................................................................................. |
Size | Perfect hit | Total Norm | Perfect Norm | SRR363963(GSM822765) AdultGlobal 5'-RACEread_length: 105. (testes) | mjTestesWT1() Testes Data. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR363960(GSM822762) Adult Library#1Small RNA Miwi IPDuplexed run:. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR363956(GSM822758) P14-WTSmall RNA Miwi IPread_length: 36. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR037903(GSM510439) testes_rep4. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|