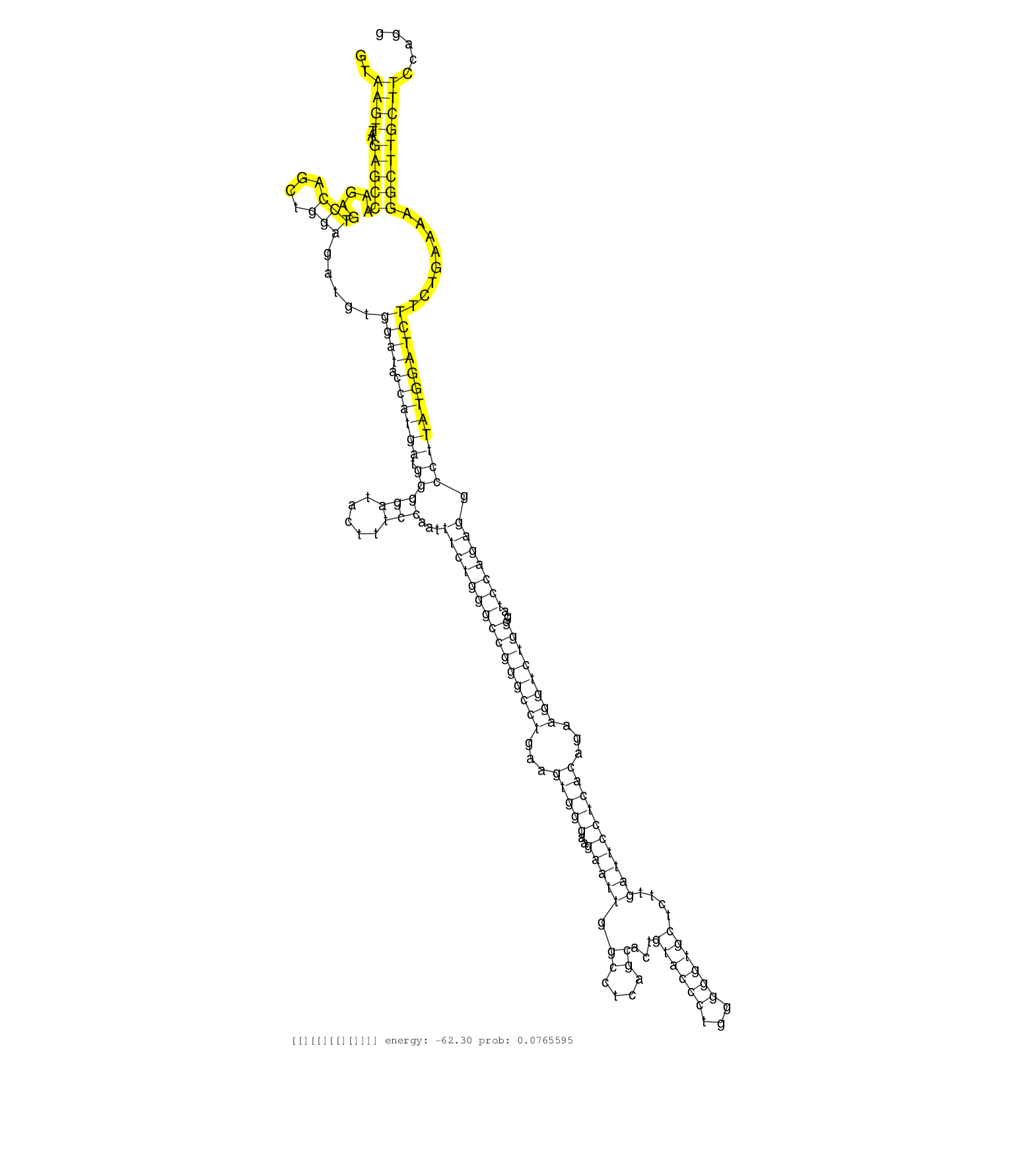

| Gene: Lass4 | ID: uc009ktx.1_intron_6_0_chr8_4520309_f | SPECIES: mm9 |
 |
 |
|
(3) OTHER.mut |
(3) PIWI.ip |
(3) PIWI.mut |
(19) TESTES |
| GCTTCTACCTTTCACTGCTGATCACTTTGCCCTTTGATGTCAAACGCAAGGTAAGTTATGAGCCAAGAGTCCAGCTGGAGATGTGGATACCATGATGGGGATACTTTCCAATTTCTGGGCCGGGCCTGAAGTGGGAAGAATTGGCCTCAGCACTGTACCCTGGGGGTGCTCTTGATTCCTCACAGAAGGTCTGGGATCCAGAGGCCTTATGGATCTTCTGAAAAGGCTTGCTTCCAGGCATGTGGCCATCCAGCCACAGAGGCTTGTCCTGGTGGTGACCCTCTGTCTTCTGCAGGACTTCAAGGAGCAGGTGGTGCACCATTTTGTGGCCGTGGGACTGATAGG ....................................................((((...(((((.....(((....))).....((((.((((((.(((((.....)))...(((((((((((((((...(((((..(((((.((....))...((((((...))))))....))))))))))...))))))))..))))))).))))))))))))........)))))))))................................................................................................................ ..................................................51.........................................................................................................................................................................................238......................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTAAGTTATGAGCCAAGAGTCCAGC.............................................................................................................................................................................................................................................................................. | 25 | 1 | 12.00 | 12.00 | 4.00 | - | 6.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGTTATGAGCCAAGAGTCCAGCT............................................................................................................................................................................................................................................................................. | 26 | 1 | 4.00 | 4.00 | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................TATGGATCTTCTGAAAAGGCTTGCTTC............................................................................................................... | 27 | 1 | 2.00 | 2.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................................................................TGGTGGTGACCCTCTGTCTTCTGCAGaa................................................ | 28 | aa | 2.00 | 0.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGTTATGAGCCAAGAGTCCAGCTG............................................................................................................................................................................................................................................................................ | 27 | 1 | 2.00 | 2.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................TACCATGATGGGGATACTTTCCAATT........................................................................................................................................................................................................................................ | 26 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ............................................................................................TGATGGGGATACTTTCCAATTTCTGGGC................................................................................................................................................................................................................................. | 28 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................TCCAGCTGGAGATGTGGATACCATGA.......................................................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ..................................................................................................................................................................................................................................................................................GTGACCCTCTGTCTTCTGCAG.................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .....................................................................................................TACTTTCCAATTTCTGGGCCGGGCC........................................................................................................................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGTTATGAGCCAAGAGTCCAGCTGGA.......................................................................................................................................................................................................................................................................... | 29 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................TGGGAAGAATTGGCCTCAGCACTGTACC.......................................................................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..............................................................................................................................TGAAGTGGGAAGAATTGGCCTCAGCAC................................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................................................................................................CTTCAAGGAGCAGGTGGTGttt.......................... | 22 | ttt | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................TGTGGATACCATGATGGGGATACTTTCC............................................................................................................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................GGAAGAATTGGCCTCAGCACTGTACCC......................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................TTCCAATTTCTGGGCCGGGCCTGAAGT..................................................................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................TGGGGATACTTTCCAATTTCTGGGCCt............................................................................................................................................................................................................................... | 27 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ............................................................................................TGATGGGGATACTTTCCAATTTCTGGGCC................................................................................................................................................................................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................TGGATCTTCTGAAAAGGCTTGCTTCCA............................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................TTCTGGGCCGGGCCTGAAGTGGGAAG............................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................CTCACAGAAGGTCTGGGATCCAGAGG............................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................TGGATCTTCTGAAAAGGCTTGCTTCC.............................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .................................................................................................................................................................................................................TGGATCTTCTGAAAAGGCTTGC.................................................................................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ............................................................................GGAGATGTGGATACCATGATG........................................................................................................................................................................................................................................................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| GCTTCTACCTTTCACTGCTGATCACTTTGCCCTTTGATGTCAAACGCAAGGTAAGTTATGAGCCAAGAGTCCAGCTGGAGATGTGGATACCATGATGGGGATACTTTCCAATTTCTGGGCCGGGCCTGAAGTGGGAAGAATTGGCCTCAGCACTGTACCCTGGGGGTGCTCTTGATTCCTCACAGAAGGTCTGGGATCCAGAGGCCTTATGGATCTTCTGAAAAGGCTTGCTTCCAGGCATGTGGCCATCCAGCCACAGAGGCTTGTCCTGGTGGTGACCCTCTGTCTTCTGCAGGACTTCAAGGAGCAGGTGGTGCACCATTTTGTGGCCGTGGGACTGATAGG ....................................................((((...(((((.....(((....))).....((((.((((((.(((((.....)))...(((((((((((((((...(((((..(((((.((....))...((((((...))))))....))))))))))...))))))))..))))))).))))))))))))........)))))))))................................................................................................................ ..................................................51.........................................................................................................................................................................................238......................................................................................................... |
Size | Perfect hit | Total Norm | Perfect Norm | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | mjTestesWT3() Testes Data. (testes) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | GSM509278(GSM509278) small RNA cloning by length. (piwi testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR028731(GSM400968) Mili-wt-associated. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR037901(GSM510437) testes_rep2. (testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................................GCCTCAGCACTGTAatag............................................................................................................................................................................................ | 18 | atag | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |