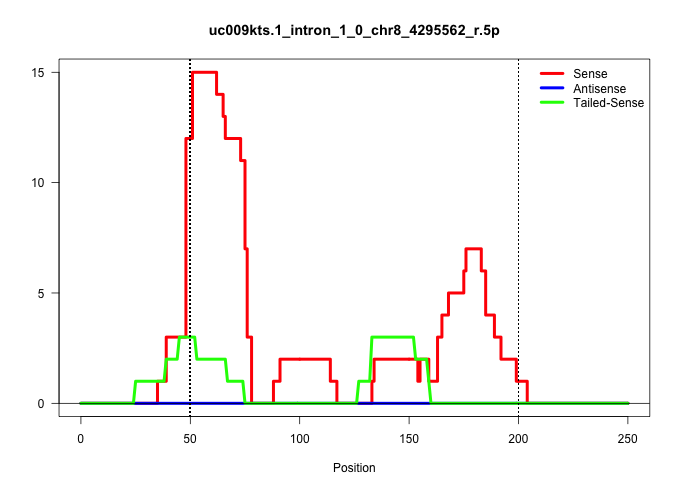
| Gene: Elavl1 | ID: uc009kts.1_intron_1_0_chr8_4295562_r.5p | SPECIES: mm9 |
 |
|
(2) OTHER.mut |
(3) PIWI.ip |
(1) PIWI.mut |
(14) TESTES |
| TCGGTTTGGGCGAATCATCAACTCCAGGGTCCTTGTGGATCAGACCACAGGTACACACAGGAGCTAAGATAGGTCTCTCTGAGCCCTAGACTAGGTGTTCTAGGGAAGATAGGGAGATATCAGCAATTAGCCTCCAAAGGAGGGCAGGTTGGACATGAGAGACCTTGGTTTTTGTTTAGTTGGGCTAGGAAAATGAAATGTCATTAAACAAATCCTGCACAACAGACAGTGGCACCTCCCTTTACTCCCA |
Size | Perfect hit | Total Norm | Perfect Norm | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................AGGTACACACAGGAGCTAAGATAGGTC............................................................................................................................................................................... | 27 | 1 | 4.00 | 4.00 | 1.00 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - |
| ................................................AGGTACACACAGGAGCTAAGATAGGTCT.............................................................................................................................................................................. | 28 | 1 | 4.00 | 4.00 | - | 1.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...................................................TACACACAGGAGCTAAGATAGGTCTCT............................................................................................................................................................................ | 27 | 1 | 3.00 | 3.00 | - | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...................................................................................................................................................................CTTGGTTTTTGTTTAGTTGGGC................................................................. | 22 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - |
| .........................AGGGTCCTTGTGGATCAGACCACAGGTt..................................................................................................................................................................................................... | 28 | t | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ................................................AGGTACACACAGGAGCTAAGATAGG................................................................................................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................GACTAGGTGTTCTAGGGAAGATAGGG........................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................CAAAGGAGGGCAGGTTGGAC................................................................................................ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .......................................TCAGACCACAGGTACACACAGGAGCTAt....................................................................................................................................................................................... | 28 | t | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................TGGTTTTTGTTTAGTTGGGCTAGGAAA.......................................................... | 27 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................CACAGGTACACACAGGAGCTAAGATAtttt............................................................................................................................................................................... | 30 | tttt | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................CCAAAGGAGGGCAGGTTGGACATGAGt.......................................................................................... | 27 | t | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................TCAGACCACAGGTACACACAGGAGCT......................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................TCAGACCACAGGTACACACAGGAGCTA........................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................TTTTTGTTTAGTTGGGCTAGG............................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...........................................................................................TAGGTGTTCTAGGGAAGATAGGGAGA..................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................CCAAAGGAGGGCAGGTTGGACATGAG........................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................CCAAAGGAGGGCAGGTTGGACATGAt........................................................................................... | 26 | t | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................TTAGTTGGGCTAGGAAAATGAAAT................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...............................................................................................................................TAGCCTCCAAAGGAGGGCAGGTTGGt................................................................................................. | 26 | t | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...........................................................................................................................................................TGAGAGACCTTGGTTTTTGTTTAGTTGG................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...................................TGGATCAGACCACAGGTACACACAGGA............................................................................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................TAGTTGGGCTAGGAAAATGAAATGTCAT.............................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| TCGGTTTGGGCGAATCATCAACTCCAGGGTCCTTGTGGATCAGACCACAGGTACACACAGGAGCTAAGATAGGTCTCTCTGAGCCCTAGACTAGGTGTTCTAGGGAAGATAGGGAGATATCAGCAATTAGCCTCCAAAGGAGGGCAGGTTGGACATGAGAGACCTTGGTTTTTGTTTAGTTGGGCTAGGAAAATGAAATGTCATTAAACAAATCCTGCACAACAGACAGTGGCACCTCCCTTTACTCCCA |
Size | Perfect hit | Total Norm | Perfect Norm | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR014235(GSM319959) 2 dpp total. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR014236(GSM319960) 10 dpp total. (testes) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | mjTestesWT2() Testes Data. (testes) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) |
|---|